Extracting Concepts for Precision Oncology from the Biomedical Literature
Paper and Code
Sep 30, 2020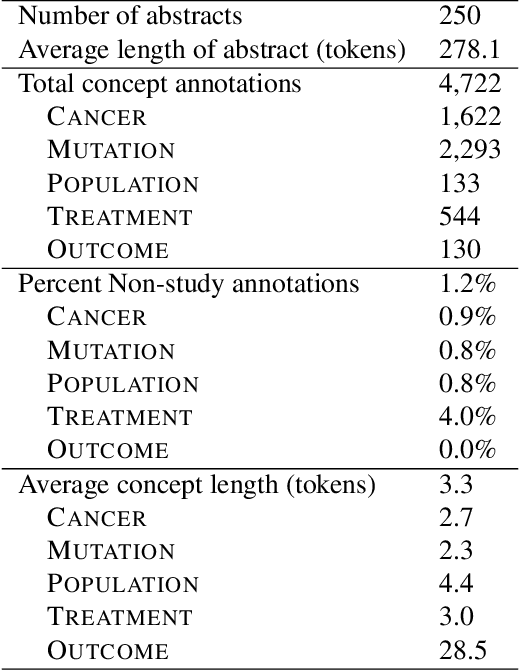
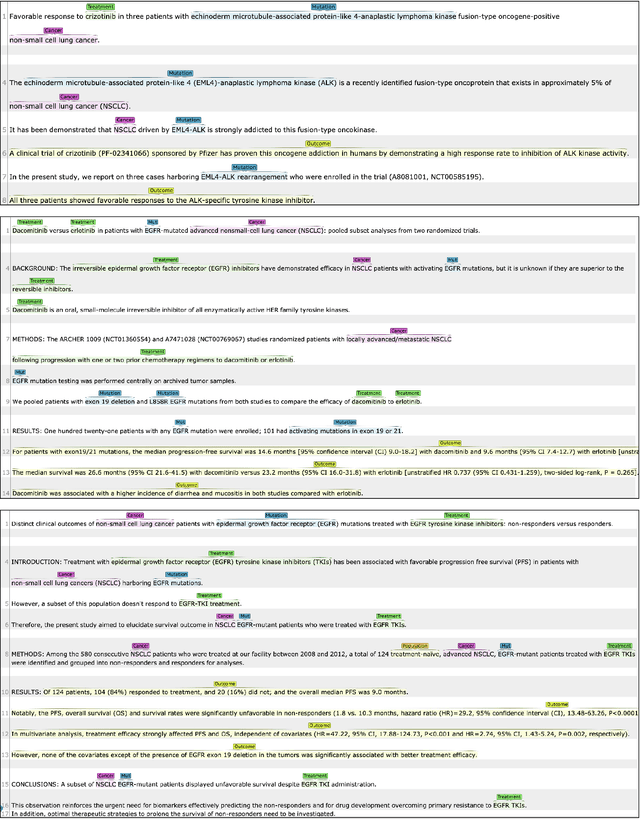

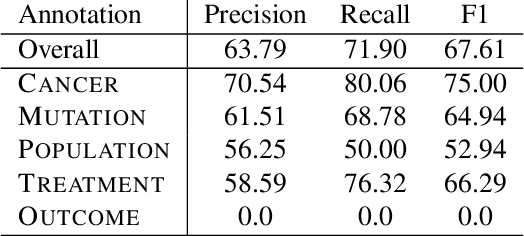
This paper describes an initial dataset and automatic natural language processing (NLP) method for extracting concepts related to precision oncology from biomedical research articles. We extract five concept types: Cancer, Mutation, Population, Treatment, Outcome. A corpus of 250 biomedical abstracts were annotated with these concepts following standard double-annotation procedures. We then experiment with BERT-based models for concept extraction. The best-performing model achieved a precision of 63.8%, a recall of 71.9%, and an F1 of 67.1. Finally, we propose additional directions for research for improving extraction performance and utilizing the NLP system in downstream precision oncology applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge