Kalpana Raja
AI-Driven Drug Repurposing through miRNA-mRNA Relation
Sep 03, 2025Abstract:miRNA mRNA relations are closely linked to several biological processes and disease mechanisms In a recent study we tested the performance of large language models LLMs on extracting miRNA mRNA relations from PubMed PubMedBERT achieved the best performance of 0.783 F1 score for miRNA mRNA Interaction Corpus MMIC Here we first applied the finetuned PubMedBERT model to extract miRNA mRNA relations from PubMed for chronic obstructive pulmonary disease COPD Alzheimers disease AD stroke type 2 diabetes mellitus T2DM chronic liver disease and cancer Next we retrieved miRNA drug relations using KinderMiner a literature mining tool for relation extraction Then we constructed three interaction networks 1 disease centric network 2 drug centric network and 3 miRNA centric network comprising 3497 nodes and 16417 edges organized as a directed graph to capture complex biological relationships Finally we validated the drugs using MIMIC IV Our integrative approach revealed both established and novel candidate drugs for diseases under study through 595 miRNA drug relations extracted from PubMed To the best of our knowledge this is the first study to systematically extract and visualize relationships among four distinct biomedical entities miRNA mRNA drug and disease
Large language models in biomedical natural language processing: benchmarks, baselines, and recommendations
May 10, 2023Abstract:Biomedical literature is growing rapidly, making it challenging to curate and extract knowledge manually. Biomedical natural language processing (BioNLP) techniques that can automatically extract information from biomedical literature help alleviate this burden. Recently, large Language Models (LLMs), such as GPT-3 and GPT-4, have gained significant attention for their impressive performance. However, their effectiveness in BioNLP tasks and impact on method development and downstream users remain understudied. This pilot study (1) establishes the baseline performance of GPT-3 and GPT-4 at both zero-shot and one-shot settings in eight BioNLP datasets across four applications: named entity recognition, relation extraction, multi-label document classification, and semantic similarity and reasoning, (2) examines the errors produced by the LLMs and categorized the errors into three types: missingness, inconsistencies, and unwanted artificial content, and (3) provides suggestions for using LLMs in BioNLP applications. We make the datasets, baselines, and results publicly available to the community via https://github.com/qingyu-qc/gpt_bionlp_benchmark.
A Hybrid Citation Retrieval Algorithm for Evidence-based Clinical Knowledge Summarization: Combining Concept Extraction, Vector Similarity and Query Expansion for High Precision
Sep 06, 2016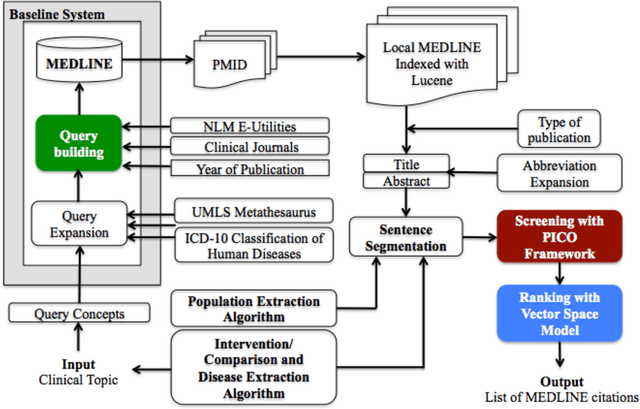
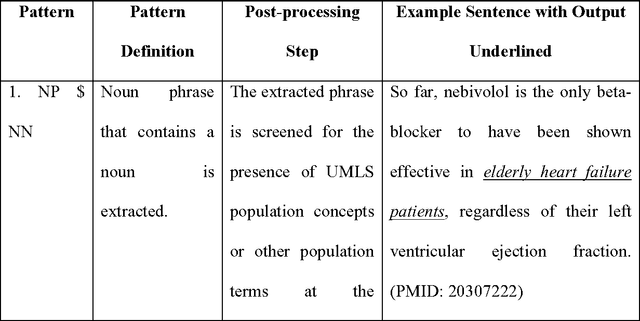
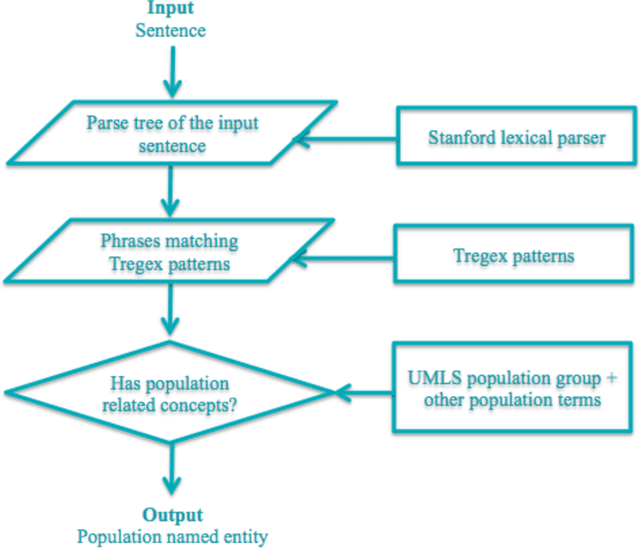

Abstract:Novel information retrieval methods to identify citations relevant to a clinical topic can overcome the knowledge gap existing between the primary literature (MEDLINE) and online clinical knowledge resources such as UpToDate. Searching the MEDLINE database directly or with query expansion methods returns a large number of citations that are not relevant to the query. The current study presents a citation retrieval system that retrieves citations for evidence-based clinical knowledge summarization. This approach combines query expansion, concept-based screening algorithm, and concept-based vector similarity. We also propose an information extraction framework for automated concept (Population, Intervention, Comparison, and Disease) extraction. We evaluated our proposed system on all topics (as queries) available from UpToDate for two diseases, heart failure (HF) and atrial fibrillation (AFib). The system achieved an overall F-score of 41.2% on HF topics and 42.4% on AFib topics on a gold standard of citations available in UpToDate. This is significantly high when compared to a query-expansion based baseline (F-score of 1.3% on HF and 2.2% on AFib) and a system that uses query expansion with disease hyponyms and journal names, concept-based screening, and term-based vector similarity system (F-score of 37.5% on HF and 39.5% on AFib). Evaluating the system with top K relevant citations, where K is the number of citations in the gold standard achieved a much higher overall F-score of 69.9% on HF topics and 75.1% on AFib topics. In addition, the system retrieved up to 18 new relevant citations per topic when tested on ten HF and six AFib clinical topics.
CRTS: A type system for representing clinical recommendations
Sep 06, 2016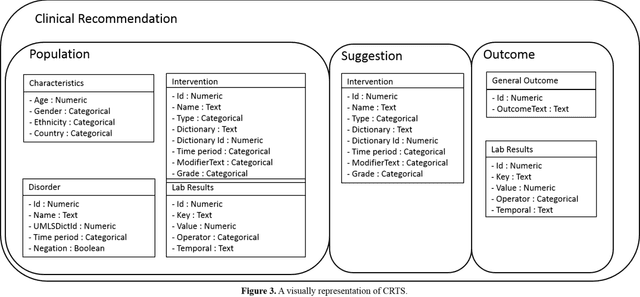
Abstract:Background: Clinical guidelines and recommendations are the driving wheels of the evidence-based medicine (EBM) paradigm, but these are available primarily as unstructured text and are generally highly heterogeneous in nature. This significantly reduces the dissemination and automatic application of these recommendations at the point of care. A comprehensive structured representation of these recommendations is highly beneficial in this regard. Objective: The objective of this paper to present Clinical Recommendation Type System (CRTS), a common type system that can effectively represent a clinical recommendation in a structured form. Methods: CRTS is built by analyzing 125 recommendations and 195 research articles corresponding to 6 different diseases available from UpToDate, a publicly available clinical knowledge system, and from the National Guideline Clearinghouse, a public resource for evidence-based clinical practice guidelines. Results: We show that CRTS not only covers the recommendations but also is flexible to be extended to represent information from primary literature. We also describe how our developed type system can be applied for clinical decision support, medical knowledge summarization, and citation retrieval. Conclusion: We showed that our proposed type system is precise and comprehensive in representing a large sample of recommendations available for various disorders. CRTS can now be used to build interoperable information extraction systems that automatically extract clinical recommendations and related data elements from clinical evidence resources, guidelines, systematic reviews and primary publications. Keywords: guidelines and recommendations, type system, clinical decision support, evidence-based medicine, information storage and retrieval
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge