Melanie R Klerer
A Hybrid Citation Retrieval Algorithm for Evidence-based Clinical Knowledge Summarization: Combining Concept Extraction, Vector Similarity and Query Expansion for High Precision
Sep 06, 2016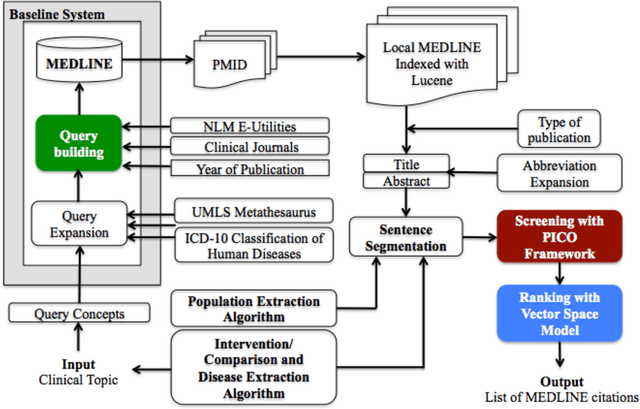
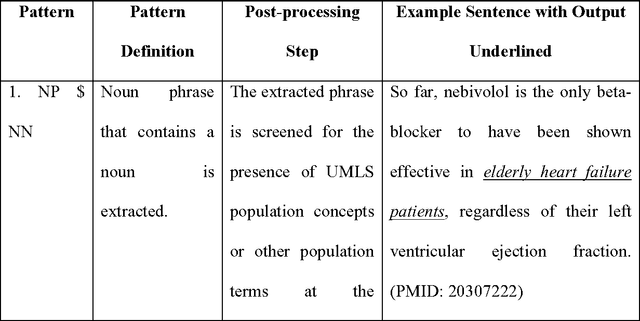
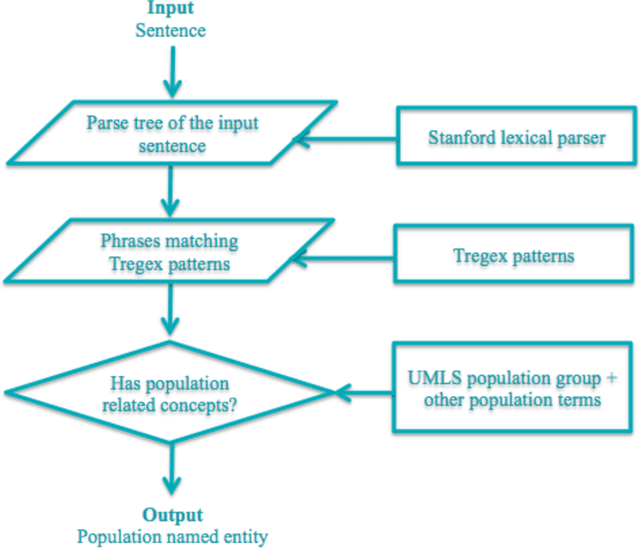

Abstract:Novel information retrieval methods to identify citations relevant to a clinical topic can overcome the knowledge gap existing between the primary literature (MEDLINE) and online clinical knowledge resources such as UpToDate. Searching the MEDLINE database directly or with query expansion methods returns a large number of citations that are not relevant to the query. The current study presents a citation retrieval system that retrieves citations for evidence-based clinical knowledge summarization. This approach combines query expansion, concept-based screening algorithm, and concept-based vector similarity. We also propose an information extraction framework for automated concept (Population, Intervention, Comparison, and Disease) extraction. We evaluated our proposed system on all topics (as queries) available from UpToDate for two diseases, heart failure (HF) and atrial fibrillation (AFib). The system achieved an overall F-score of 41.2% on HF topics and 42.4% on AFib topics on a gold standard of citations available in UpToDate. This is significantly high when compared to a query-expansion based baseline (F-score of 1.3% on HF and 2.2% on AFib) and a system that uses query expansion with disease hyponyms and journal names, concept-based screening, and term-based vector similarity system (F-score of 37.5% on HF and 39.5% on AFib). Evaluating the system with top K relevant citations, where K is the number of citations in the gold standard achieved a much higher overall F-score of 69.9% on HF topics and 75.1% on AFib topics. In addition, the system retrieved up to 18 new relevant citations per topic when tested on ten HF and six AFib clinical topics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge