Tianhao Li
additional authors not shown
Emerging from Ground: Addressing Intent Deviation in Tool-Using Agents via Deriving Real Calls into Virtual Trajectories
Jan 21, 2026Abstract:LLMs have advanced tool-using agents for real-world applications, yet they often lead to unexpected behaviors or results. Beyond obvious failures, the subtle issue of "intent deviation" severely hinders reliable evaluation and performance improvement. Existing post-training methods generally leverage either real system samples or virtual data simulated by LLMs. However, the former is costly due to reliance on hand-crafted user requests, while the latter suffers from distribution shift from the real tools in the wild. Additionally, both methods lack negative samples tailored to intent deviation scenarios, hindering effective guidance on preference learning. We introduce RISE, a "Real-to-Virtual" method designed to mitigate intent deviation. Anchoring on verified tool primitives, RISE synthesizes virtual trajectories and generates diverse negative samples through mutation on critical parameters. With synthetic data, RISE fine-tunes backbone LLMs via the two-stage training for intent alignment. Evaluation results demonstrate that data synthesized by RISE achieve promising results in eight metrics covering user requires, execution trajectories and agent responses. Integrating with training, RISE achieves an average 35.28% improvement in Acctask (task completion) and 23.27% in Accintent (intent alignment), outperforming SOTA baselines by 1.20--42.09% and 1.17--54.93% respectively.
AI Benchmark Democratization and Carpentry
Dec 12, 2025Abstract:Benchmarks are a cornerstone of modern machine learning, enabling reproducibility, comparison, and scientific progress. However, AI benchmarks are increasingly complex, requiring dynamic, AI-focused workflows. Rapid evolution in model architectures, scale, datasets, and deployment contexts makes evaluation a moving target. Large language models often memorize static benchmarks, causing a gap between benchmark results and real-world performance. Beyond traditional static benchmarks, continuous adaptive benchmarking frameworks are needed to align scientific assessment with deployment risks. This calls for skills and education in AI Benchmark Carpentry. From our experience with MLCommons, educational initiatives, and programs like the DOE's Trillion Parameter Consortium, key barriers include high resource demands, limited access to specialized hardware, lack of benchmark design expertise, and uncertainty in relating results to application domains. Current benchmarks often emphasize peak performance on top-tier hardware, offering limited guidance for diverse, real-world scenarios. Benchmarking must become dynamic, incorporating evolving models, updated data, and heterogeneous platforms while maintaining transparency, reproducibility, and interpretability. Democratization requires both technical innovation and systematic education across levels, building sustained expertise in benchmark design and use. Benchmarks should support application-relevant comparisons, enabling informed, context-sensitive decisions. Dynamic, inclusive benchmarking will ensure evaluation keeps pace with AI evolution and supports responsible, reproducible, and accessible AI deployment. Community efforts can provide a foundation for AI Benchmark Carpentry.
A Survey of Vibe Coding with Large Language Models
Oct 14, 2025Abstract:The advancement of large language models (LLMs) has catalyzed a paradigm shift from code generation assistance to autonomous coding agents, enabling a novel development methodology termed "Vibe Coding" where developers validate AI-generated implementations through outcome observation rather than line-by-line code comprehension. Despite its transformative potential, the effectiveness of this emergent paradigm remains under-explored, with empirical evidence revealing unexpected productivity losses and fundamental challenges in human-AI collaboration. To address this gap, this survey provides the first comprehensive and systematic review of Vibe Coding with large language models, establishing both theoretical foundations and practical frameworks for this transformative development approach. Drawing from systematic analysis of over 1000 research papers, we survey the entire vibe coding ecosystem, examining critical infrastructure components including LLMs for coding, LLM-based coding agent, development environment of coding agent, and feedback mechanisms. We first introduce Vibe Coding as a formal discipline by formalizing it through a Constrained Markov Decision Process that captures the dynamic triadic relationship among human developers, software projects, and coding agents. Building upon this theoretical foundation, we then synthesize existing practices into five distinct development models: Unconstrained Automation, Iterative Conversational Collaboration, Planning-Driven, Test-Driven, and Context-Enhanced Models, thus providing the first comprehensive taxonomy in this domain. Critically, our analysis reveals that successful Vibe Coding depends not merely on agent capabilities but on systematic context engineering, well-established development environments, and human-agent collaborative development models.
MMOT: The First Challenging Benchmark for Drone-based Multispectral Multi-Object Tracking
Oct 14, 2025Abstract:Drone-based multi-object tracking is essential yet highly challenging due to small targets, severe occlusions, and cluttered backgrounds. Existing RGB-based tracking algorithms heavily depend on spatial appearance cues such as color and texture, which often degrade in aerial views, compromising reliability. Multispectral imagery, capturing pixel-level spectral reflectance, provides crucial cues that enhance object discriminability under degraded spatial conditions. However, the lack of dedicated multispectral UAV datasets has hindered progress in this domain. To bridge this gap, we introduce MMOT, the first challenging benchmark for drone-based multispectral multi-object tracking. It features three key characteristics: (i) Large Scale - 125 video sequences with over 488.8K annotations across eight categories; (ii) Comprehensive Challenges - covering diverse conditions such as extreme small targets, high-density scenarios, severe occlusions, and complex motion; and (iii) Precise Oriented Annotations - enabling accurate localization and reduced ambiguity under aerial perspectives. To better extract spectral features and leverage oriented annotations, we further present a multispectral and orientation-aware MOT scheme adapting existing methods, featuring: (i) a lightweight Spectral 3D-Stem integrating spectral features while preserving compatibility with RGB pretraining; (ii) an orientation-aware Kalman filter for precise state estimation; and (iii) an end-to-end orientation-adaptive transformer. Extensive experiments across representative trackers consistently show that multispectral input markedly improves tracking performance over RGB baselines, particularly for small and densely packed objects. We believe our work will advance drone-based multispectral multi-object tracking research. Our MMOT, code, and benchmarks are publicly available at https://github.com/Annzstbl/MMOT.
GTAD: Global Temporal Aggregation Denoising Learning for 3D Semantic Occupancy Prediction
Jul 28, 2025Abstract:Accurately perceiving dynamic environments is a fundamental task for autonomous driving and robotic systems. Existing methods inadequately utilize temporal information, relying mainly on local temporal interactions between adjacent frames and failing to leverage global sequence information effectively. To address this limitation, we investigate how to effectively aggregate global temporal features from temporal sequences, aiming to achieve occupancy representations that efficiently utilize global temporal information from historical observations. For this purpose, we propose a global temporal aggregation denoising network named GTAD, introducing a global temporal information aggregation framework as a new paradigm for holistic 3D scene understanding. Our method employs an in-model latent denoising network to aggregate local temporal features from the current moment and global temporal features from historical sequences. This approach enables the effective perception of both fine-grained temporal information from adjacent frames and global temporal patterns from historical observations. As a result, it provides a more coherent and comprehensive understanding of the environment. Extensive experiments on the nuScenes and Occ3D-nuScenes benchmark and ablation studies demonstrate the superiority of our method.
Qwen3 Technical Report
May 14, 2025
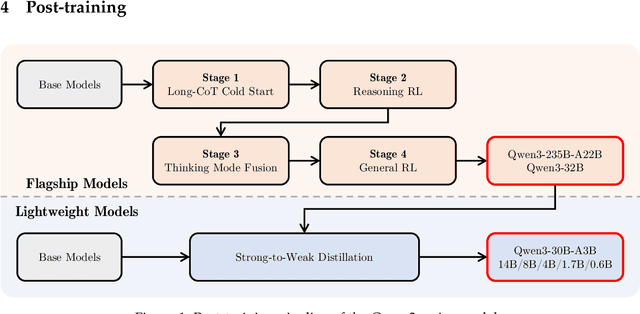
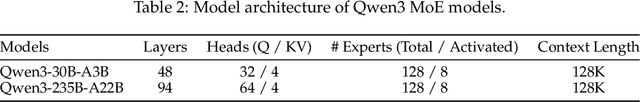
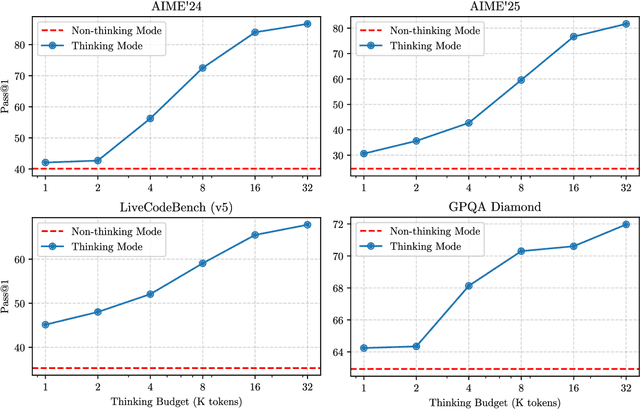
Abstract:In this work, we present Qwen3, the latest version of the Qwen model family. Qwen3 comprises a series of large language models (LLMs) designed to advance performance, efficiency, and multilingual capabilities. The Qwen3 series includes models of both dense and Mixture-of-Expert (MoE) architectures, with parameter scales ranging from 0.6 to 235 billion. A key innovation in Qwen3 is the integration of thinking mode (for complex, multi-step reasoning) and non-thinking mode (for rapid, context-driven responses) into a unified framework. This eliminates the need to switch between different models--such as chat-optimized models (e.g., GPT-4o) and dedicated reasoning models (e.g., QwQ-32B)--and enables dynamic mode switching based on user queries or chat templates. Meanwhile, Qwen3 introduces a thinking budget mechanism, allowing users to allocate computational resources adaptively during inference, thereby balancing latency and performance based on task complexity. Moreover, by leveraging the knowledge from the flagship models, we significantly reduce the computational resources required to build smaller-scale models, while ensuring their highly competitive performance. Empirical evaluations demonstrate that Qwen3 achieves state-of-the-art results across diverse benchmarks, including tasks in code generation, mathematical reasoning, agent tasks, etc., competitive against larger MoE models and proprietary models. Compared to its predecessor Qwen2.5, Qwen3 expands multilingual support from 29 to 119 languages and dialects, enhancing global accessibility through improved cross-lingual understanding and generation capabilities. To facilitate reproducibility and community-driven research and development, all Qwen3 models are publicly accessible under Apache 2.0.
Enhancing LLM Language Adaption through Cross-lingual In-Context Pre-training
Apr 29, 2025Abstract:Large language models (LLMs) exhibit remarkable multilingual capabilities despite English-dominated pre-training, attributed to cross-lingual mechanisms during pre-training. Existing methods for enhancing cross-lingual transfer remain constrained by parallel resources, suffering from limited linguistic and domain coverage. We propose Cross-lingual In-context Pre-training (CrossIC-PT), a simple and scalable approach that enhances cross-lingual transfer by leveraging semantically related bilingual texts via simple next-word prediction. We construct CrossIC-PT samples by interleaving semantic-related bilingual Wikipedia documents into a single context window. To access window size constraints, we implement a systematic segmentation policy to split long bilingual document pairs into chunks while adjusting the sliding window mechanism to preserve contextual coherence. We further extend data availability through a semantic retrieval framework to construct CrossIC-PT samples from web-crawled corpus. Experimental results demonstrate that CrossIC-PT improves multilingual performance on three models (Llama-3.1-8B, Qwen2.5-7B, and Qwen2.5-1.5B) across six target languages, yielding performance gains of 3.79%, 3.99%, and 1.95%, respectively, with additional improvements after data augmentation.
GuideLLM: Exploring LLM-Guided Conversation with Applications in Autobiography Interviewing
Feb 10, 2025



Abstract:Although Large Language Models (LLMs) succeed in human-guided conversations such as instruction following and question answering, the potential of LLM-guided conversations-where LLMs direct the discourse and steer the conversation's objectives-remains under-explored. In this study, we first characterize LLM-guided conversation into three fundamental components: (i) Goal Navigation; (ii) Context Management; (iii) Empathetic Engagement, and propose GuideLLM as an installation. We then implement an interviewing environment for the evaluation of LLM-guided conversation. Specifically, various topics are involved in this environment for comprehensive interviewing evaluation, resulting in around 1.4k turns of utterances, 184k tokens, and over 200 events mentioned during the interviewing for each chatbot evaluation. We compare GuideLLM with 6 state-of-the-art LLMs such as GPT-4o and Llama-3-70b-Instruct, from the perspective of interviewing quality, and autobiography generation quality. For automatic evaluation, we derive user proxies from multiple autobiographies and employ LLM-as-a-judge to score LLM behaviors. We further conduct a human-involved experiment by employing 45 human participants to chat with GuideLLM and baselines. We then collect human feedback, preferences, and ratings regarding the qualities of conversation and autobiography. Experimental results indicate that GuideLLM significantly outperforms baseline LLMs in automatic evaluation and achieves consistent leading performances in human ratings.
Synthetic Poisoning Attacks: The Impact of Poisoned MRI Image on U-Net Brain Tumor Segmentation
Feb 06, 2025Abstract:Deep learning-based medical image segmentation models, such as U-Net, rely on high-quality annotated datasets to achieve accurate predictions. However, the increasing use of generative models for synthetic data augmentation introduces potential risks, particularly in the absence of rigorous quality control. In this paper, we investigate the impact of synthetic MRI data on the robustness and segmentation accuracy of U-Net models for brain tumor segmentation. Specifically, we generate synthetic T1-contrast-enhanced (T1-Ce) MRI scans using a GAN-based model with a shared encoding-decoding framework and shortest-path regularization. To quantify the effect of synthetic data contamination, we train U-Net models on progressively "poisoned" datasets, where synthetic data proportions range from 16.67% to 83.33%. Experimental results on a real MRI validation set reveal a significant performance degradation as synthetic data increases, with Dice coefficients dropping from 0.8937 (33.33% synthetic) to 0.7474 (83.33% synthetic). Accuracy and sensitivity exhibit similar downward trends, demonstrating the detrimental effect of synthetic data on segmentation robustness. These findings underscore the importance of quality control in synthetic data integration and highlight the risks of unregulated synthetic augmentation in medical image analysis. Our study provides critical insights for the development of more reliable and trustworthy AI-driven medical imaging systems.
Qwen2.5 Technical Report
Dec 19, 2024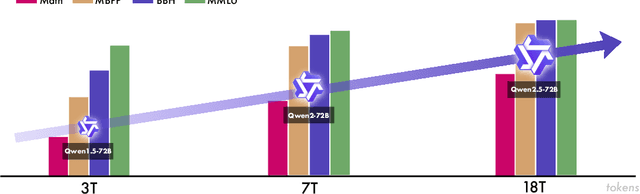

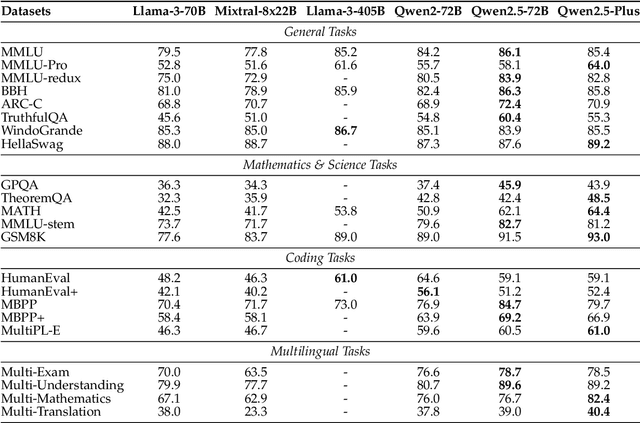
Abstract:In this report, we introduce Qwen2.5, a comprehensive series of large language models (LLMs) designed to meet diverse needs. Compared to previous iterations, Qwen 2.5 has been significantly improved during both the pre-training and post-training stages. In terms of pre-training, we have scaled the high-quality pre-training datasets from the previous 7 trillion tokens to 18 trillion tokens. This provides a strong foundation for common sense, expert knowledge, and reasoning capabilities. In terms of post-training, we implement intricate supervised finetuning with over 1 million samples, as well as multistage reinforcement learning. Post-training techniques enhance human preference, and notably improve long text generation, structural data analysis, and instruction following. To handle diverse and varied use cases effectively, we present Qwen2.5 LLM series in rich sizes. Open-weight offerings include base and instruction-tuned models, with quantized versions available. In addition, for hosted solutions, the proprietary models currently include two mixture-of-experts (MoE) variants: Qwen2.5-Turbo and Qwen2.5-Plus, both available from Alibaba Cloud Model Studio. Qwen2.5 has demonstrated top-tier performance on a wide range of benchmarks evaluating language understanding, reasoning, mathematics, coding, human preference alignment, etc. Specifically, the open-weight flagship Qwen2.5-72B-Instruct outperforms a number of open and proprietary models and demonstrates competitive performance to the state-of-the-art open-weight model, Llama-3-405B-Instruct, which is around 5 times larger. Qwen2.5-Turbo and Qwen2.5-Plus offer superior cost-effectiveness while performing competitively against GPT-4o-mini and GPT-4o respectively. Additionally, as the foundation, Qwen2.5 models have been instrumental in training specialized models such as Qwen2.5-Math, Qwen2.5-Coder, QwQ, and multimodal models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge