Tianyu Zeng
Synthetic Poisoning Attacks: The Impact of Poisoned MRI Image on U-Net Brain Tumor Segmentation
Feb 06, 2025Abstract:Deep learning-based medical image segmentation models, such as U-Net, rely on high-quality annotated datasets to achieve accurate predictions. However, the increasing use of generative models for synthetic data augmentation introduces potential risks, particularly in the absence of rigorous quality control. In this paper, we investigate the impact of synthetic MRI data on the robustness and segmentation accuracy of U-Net models for brain tumor segmentation. Specifically, we generate synthetic T1-contrast-enhanced (T1-Ce) MRI scans using a GAN-based model with a shared encoding-decoding framework and shortest-path regularization. To quantify the effect of synthetic data contamination, we train U-Net models on progressively "poisoned" datasets, where synthetic data proportions range from 16.67% to 83.33%. Experimental results on a real MRI validation set reveal a significant performance degradation as synthetic data increases, with Dice coefficients dropping from 0.8937 (33.33% synthetic) to 0.7474 (83.33% synthetic). Accuracy and sensitivity exhibit similar downward trends, demonstrating the detrimental effect of synthetic data on segmentation robustness. These findings underscore the importance of quality control in synthetic data integration and highlight the risks of unregulated synthetic augmentation in medical image analysis. Our study provides critical insights for the development of more reliable and trustworthy AI-driven medical imaging systems.
SciSafeEval: A Comprehensive Benchmark for Safety Alignment of Large Language Models in Scientific Tasks
Oct 02, 2024



Abstract:Large language models (LLMs) have had a transformative impact on a variety of scientific tasks across disciplines such as biology, chemistry, medicine, and physics. However, ensuring the safety alignment of these models in scientific research remains an underexplored area, with existing benchmarks primarily focus on textual content and overlooking key scientific representations such as molecular, protein, and genomic languages. Moreover, the safety mechanisms of LLMs in scientific tasks are insufficiently studied. To address these limitations, we introduce SciSafeEval, a comprehensive benchmark designed to evaluate the safety alignment of LLMs across a range of scientific tasks. SciSafeEval spans multiple scientific languages - including textual, molecular, protein, and genomic - and covers a wide range of scientific domains. We evaluate LLMs in zero-shot, few-shot and chain-of-thought settings, and introduce a 'jailbreak' enhancement feature that challenges LLMs equipped with safety guardrails, rigorously testing their defenses against malicious intention. Our benchmark surpasses existing safety datasets in both scale and scope, providing a robust platform for assessing the safety and performance of LLMs in scientific contexts. This work aims to facilitate the responsible development and deployment of LLMs, promoting alignment with safety and ethical standards in scientific research.
A Sub-pixel Accurate Quantification of Joint Space Narrowing Progression in Rheumatoid Arthritis
May 19, 2022
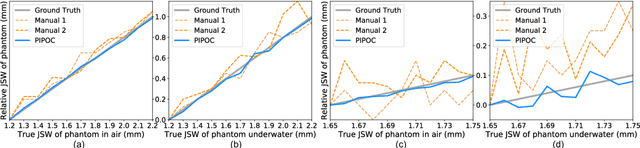
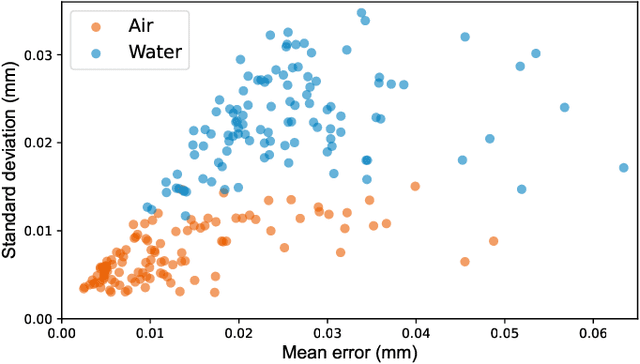
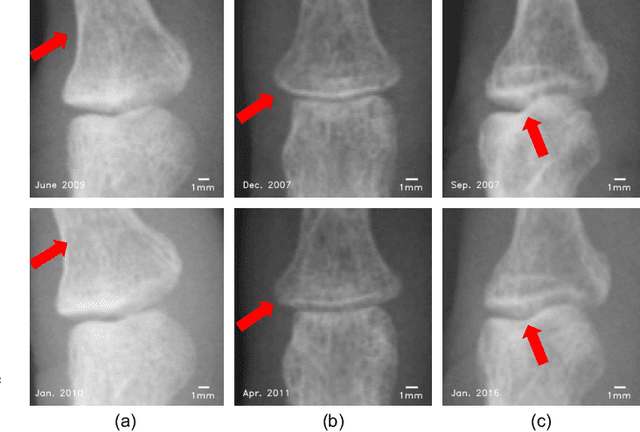
Abstract:Rheumatoid arthritis (RA) is a chronic autoimmune disease that primarily affects peripheral synovial joints, like fingers, wrist and feet. Radiology plays a critical role in the diagnosis and monitoring of RA. Limited by the current spatial resolution of radiographic imaging, joint space narrowing (JSN) progression of RA with the same reason above can be less than one pixel per year with universal spatial resolution. Insensitive monitoring of JSN can hinder the radiologist/rheumatologist from making a proper and timely clinical judgment. In this paper, we propose a novel and sensitive method that we call partial image phase-only correlation which aims to automatically quantify JSN progression in the early stages of RA. The majority of the current literature utilizes the mean error, root-mean-square deviation and standard deviation to report the accuracy at pixel level. Our work measures JSN progression between a baseline and its follow-up finger joint images by using the phase spectrum in the frequency domain. Using this study, the mean error can be reduced to 0.0130mm when applied to phantom radiographs with ground truth, and 0.0519mm standard deviation for clinical radiography. With its sub-pixel accuracy far beyond manual measurement, we are optimistic that our work is promising for automatically quantifying JSN progression.
Predictions of 2019-nCoV Transmission Ending via Comprehensive Methods
Feb 20, 2020
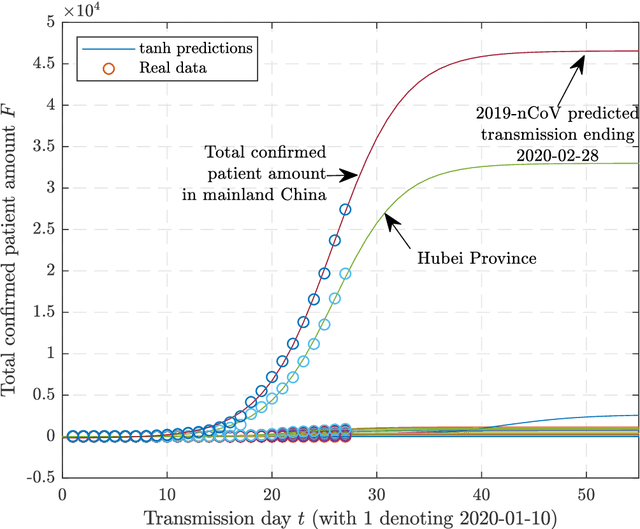

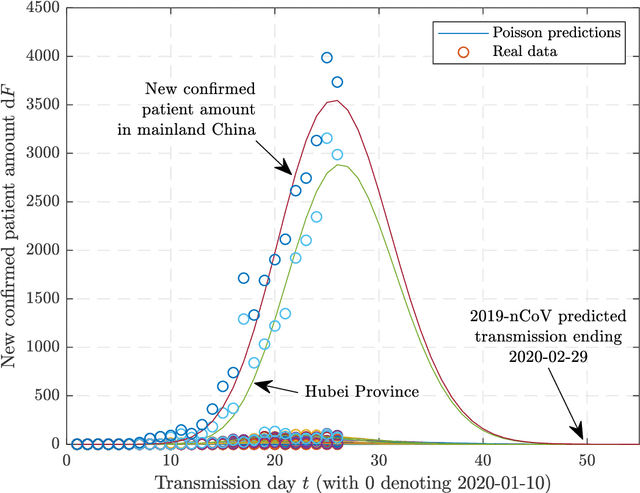
Abstract:Since the SARS outbreak in 2003, a lot of predictive epidemiological models have been proposed. At the end of 2019, a novel coronavirus, termed as 2019-nCoV, has broken out and is propagating in China and the world. Here we propose a multi-model ordinary differential equation set neural network (MMODEs-NN) and model-free methods to predict the interprovincial transmissions in mainland China, especially those from Hubei Province. Compared with the previously proposed epidemiological models, the proposed network can simulate the transportations with the ODEs activation method, while the model-free methods based on the sigmoid function, Gaussian function, and Poisson distribution are linear and fast to generate reasonable predictions. According to the numerical experiments and the realities, the special policies for controlling the disease are successful in some provinces, and the transmission of the epidemic, whose outbreak time is close to the beginning of China Spring Festival travel rush, is more likely to decelerate before February 18 and to end before April 2020. The proposed mathematical and artificial intelligence methods can give consistent and reasonable predictions of the 2019-nCoV ending. We anticipate our work to be a starting point for comprehensive prediction researches of the 2019-nCoV.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge