Thomas C. Shen
Universal Lymph Node Detection in Multiparametric MRI with Selective Augmentation
Apr 07, 2025Abstract:Robust localization of lymph nodes (LNs) in multiparametric MRI (mpMRI) is critical for the assessment of lymphadenopathy. Radiologists routinely measure the size of LN to distinguish benign from malignant nodes, which would require subsequent cancer staging. Sizing is a cumbersome task compounded by the diverse appearances of LNs in mpMRI, which renders their measurement difficult. Furthermore, smaller and potentially metastatic LNs could be missed during a busy clinical day. To alleviate these imaging and workflow problems, we propose a pipeline to universally detect both benign and metastatic nodes in the body for their ensuing measurement. The recently proposed VFNet neural network was employed to identify LN in T2 fat suppressed and diffusion weighted imaging (DWI) sequences acquired by various scanners with a variety of exam protocols. We also use a selective augmentation technique known as Intra-Label LISA (ILL) to diversify the input data samples the model sees during training, such that it improves its robustness during the evaluation phase. We achieved a sensitivity of $\sim$83\% with ILL vs. $\sim$80\% without ILL at 4 FP/vol. Compared with current LN detection approaches evaluated on mpMRI, we show a sensitivity improvement of $\sim$9\% at 4 FP/vol.
How Does Pruning Impact Long-Tailed Multi-Label Medical Image Classifiers?
Aug 17, 2023Abstract:Pruning has emerged as a powerful technique for compressing deep neural networks, reducing memory usage and inference time without significantly affecting overall performance. However, the nuanced ways in which pruning impacts model behavior are not well understood, particularly for long-tailed, multi-label datasets commonly found in clinical settings. This knowledge gap could have dangerous implications when deploying a pruned model for diagnosis, where unexpected model behavior could impact patient well-being. To fill this gap, we perform the first analysis of pruning's effect on neural networks trained to diagnose thorax diseases from chest X-rays (CXRs). On two large CXR datasets, we examine which diseases are most affected by pruning and characterize class "forgettability" based on disease frequency and co-occurrence behavior. Further, we identify individual CXRs where uncompressed and heavily pruned models disagree, known as pruning-identified exemplars (PIEs), and conduct a human reader study to evaluate their unifying qualities. We find that radiologists perceive PIEs as having more label noise, lower image quality, and higher diagnosis difficulty. This work represents a first step toward understanding the impact of pruning on model behavior in deep long-tailed, multi-label medical image classification. All code, model weights, and data access instructions can be found at https://github.com/VITA-Group/PruneCXR.
Improving Segmentation and Detection of Lesions in CT Scans Using Intensity Distribution Supervision
Jul 11, 2023Abstract:We propose a method to incorporate the intensity information of a target lesion on CT scans in training segmentation and detection networks. We first build an intensity-based lesion probability (ILP) function from an intensity histogram of the target lesion. It is used to compute the probability of being the lesion for each voxel based on its intensity. Finally, the computed ILP map of each input CT scan is provided as additional supervision for network training, which aims to inform the network about possible lesion locations in terms of intensity values at no additional labeling cost. The method was applied to improve the segmentation of three different lesion types, namely, small bowel carcinoid tumor, kidney tumor, and lung nodule. The effectiveness of the proposed method on a detection task was also investigated. We observed improvements of 41.3% -> 47.8%, 74.2% -> 76.0%, and 26.4% -> 32.7% in segmenting small bowel carcinoid tumor, kidney tumor, and lung nodule, respectively, in terms of per case Dice scores. An improvement of 64.6% -> 75.5% was achieved in detecting kidney tumors in terms of average precision. The results of different usages of the ILP map and the effect of varied amount of training data are also presented.
Long-Tailed Classification of Thorax Diseases on Chest X-Ray: A New Benchmark Study
Aug 29, 2022

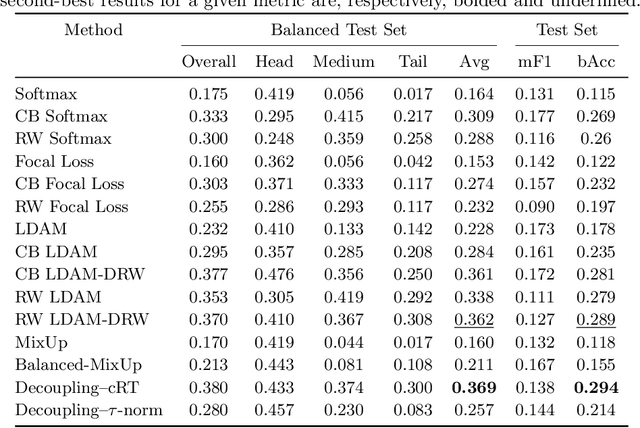

Abstract:Imaging exams, such as chest radiography, will yield a small set of common findings and a much larger set of uncommon findings. While a trained radiologist can learn the visual presentation of rare conditions by studying a few representative examples, teaching a machine to learn from such a "long-tailed" distribution is much more difficult, as standard methods would be easily biased toward the most frequent classes. In this paper, we present a comprehensive benchmark study of the long-tailed learning problem in the specific domain of thorax diseases on chest X-rays. We focus on learning from naturally distributed chest X-ray data, optimizing classification accuracy over not only the common "head" classes, but also the rare yet critical "tail" classes. To accomplish this, we introduce a challenging new long-tailed chest X-ray benchmark to facilitate research on developing long-tailed learning methods for medical image classification. The benchmark consists of two chest X-ray datasets for 19- and 20-way thorax disease classification, containing classes with as many as 53,000 and as few as 7 labeled training images. We evaluate both standard and state-of-the-art long-tailed learning methods on this new benchmark, analyzing which aspects of these methods are most beneficial for long-tailed medical image classification and summarizing insights for future algorithm design. The datasets, trained models, and code are available at https://github.com/VITA-Group/LongTailCXR.
Improving Small Lesion Segmentation in CT Scans using Intensity Distribution Supervision: Application to Small Bowel Carcinoid Tumor
Jul 29, 2022
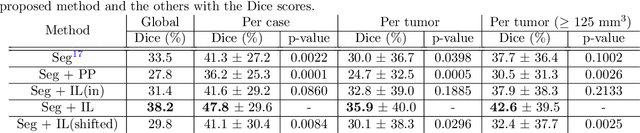
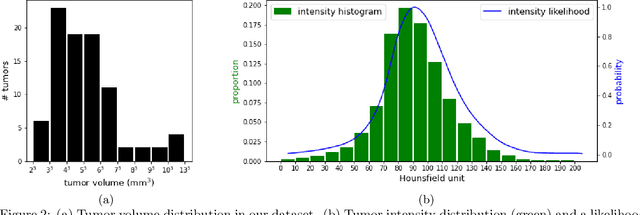
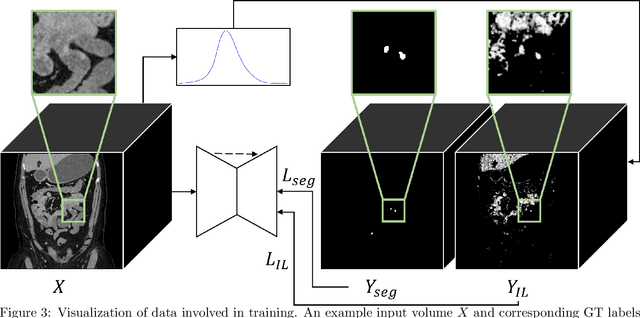
Abstract:Finding small lesions is very challenging due to lack of noticeable features, severe class imbalance, as well as the size itself. One approach to improve small lesion segmentation is to reduce the region of interest and inspect it at a higher sensitivity rather than performing it for the entire region. It is usually implemented as sequential or joint segmentation of organ and lesion, which requires additional supervision on organ segmentation. Instead, we propose to utilize an intensity distribution of a target lesion at no additional labeling cost to effectively separate regions where the lesions are possibly located from the background. It is incorporated into network training as an auxiliary task. We applied the proposed method to segmentation of small bowel carcinoid tumors in CT scans. We observed improvements for all metrics (33.5% $\rightarrow$ 38.2%, 41.3% $\rightarrow$ 47.8%, 30.0% $\rightarrow$ 35.9% for the global, per case, and per tumor Dice scores, respectively.) compared to the baseline method, which proves the validity of our idea. Our method can be one option for explicitly incorporating intensity distribution information of a target in network training.
Universal Lymph Node Detection in T2 MRI using Neural Networks
Mar 31, 2022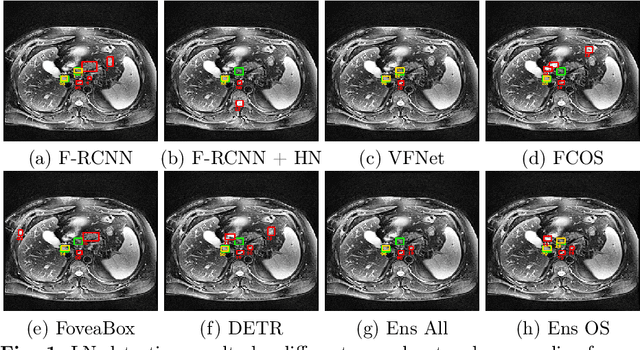
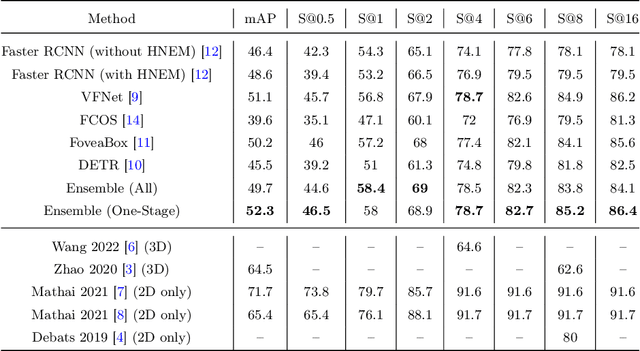
Abstract:Purpose: Identification of abdominal Lymph Nodes (LN) that are suspicious for metastasis in T2 Magnetic Resonance Imaging (MRI) scans is critical for staging of lymphoproliferative diseases. Prior work on LN detection has been limited to specific anatomical regions of the body (pelvis, rectum) in single MR slices. Therefore, the development of a universal approach to detect LN in full T2 MRI volumes is highly desirable. Methods: In this study, a Computer Aided Detection (CAD) pipeline to universally identify abdominal LN in volumetric T2 MRI using neural networks is proposed. First, we trained various neural network models for detecting LN: Faster RCNN with and without Hard Negative Example Mining (HNEM), FCOS, FoveaBox, VFNet, and Detection Transformer (DETR). Next, we show that the state-of-the-art (SOTA) VFNet model with Adaptive Training Sample Selection (ATSS) outperforms Faster RCNN with HNEM. Finally, we ensembled models that surpassed a 45% mAP threshold. We found that the VFNet model and one-stage model ensemble can be interchangeably used in the CAD pipeline. Results: Experiments on 122 test T2 MRI volumes revealed that VFNet achieved a 51.1% mAP and 78.7% recall at 4 false positives (FP) per volume, while the one-stage model ensemble achieved a mAP of 52.3% and sensitivity of 78.7% at 4FP. Conclusion: Our contribution is a CAD pipeline that detects LN in T2 MRI volumes, resulting in a sensitivity improvement of $\sim$14 points over the current SOTA method for LN detection (sensitivity of 78.7% at 4 FP vs. 64.6% at 5 FP per volume).
Lymph Node Detection in T2 MRI with Transformers
Nov 09, 2021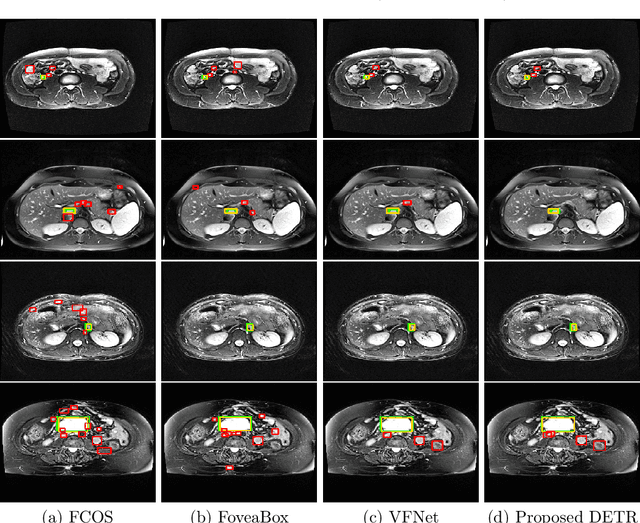
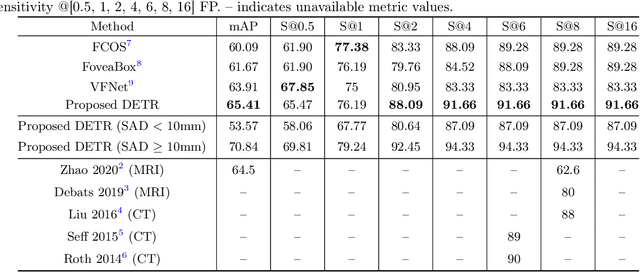
Abstract:Identification of lymph nodes (LN) in T2 Magnetic Resonance Imaging (MRI) is an important step performed by radiologists during the assessment of lymphoproliferative diseases. The size of the nodes play a crucial role in their staging, and radiologists sometimes use an additional contrast sequence such as diffusion weighted imaging (DWI) for confirmation. However, lymph nodes have diverse appearances in T2 MRI scans, making it tough to stage for metastasis. Furthermore, radiologists often miss smaller metastatic lymph nodes over the course of a busy day. To deal with these issues, we propose to use the DEtection TRansformer (DETR) network to localize suspicious metastatic lymph nodes for staging in challenging T2 MRI scans acquired by different scanners and exam protocols. False positives (FP) were reduced through a bounding box fusion technique, and a precision of 65.41\% and sensitivity of 91.66\% at 4 FP per image was achieved. To the best of our knowledge, our results improve upon the current state-of-the-art for lymph node detection in T2 MRI scans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge