Perry J. Pickhardt
Utility of Pancreas Surface Lobularity as a CT Biomarker for Opportunistic Screening of Type 2 Diabetes
Nov 13, 2025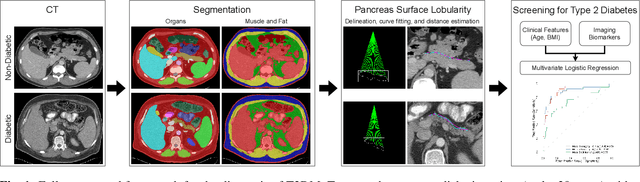
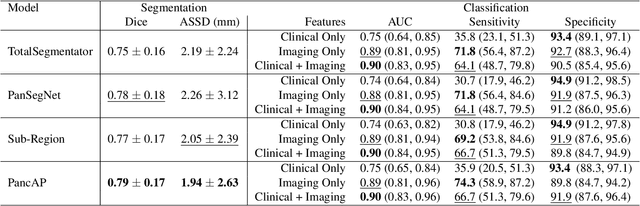
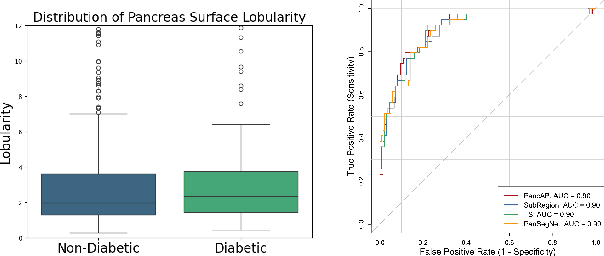
Abstract:Type 2 Diabetes Mellitus (T2DM) is a chronic metabolic disease that affects millions of people worldwide. Early detection is crucial as it can alter pancreas function through morphological changes and increased deposition of ectopic fat, eventually leading to organ damage. While studies have shown an association between T2DM and pancreas volume and fat content, the role of increased pancreatic surface lobularity (PSL) in patients with T2DM has not been fully investigated. In this pilot work, we propose a fully automated approach to delineate the pancreas and other abdominal structures, derive CT imaging biomarkers, and opportunistically screen for T2DM. Four deep learning-based models were used to segment the pancreas in an internal dataset of 584 patients (297 males, 437 non-diabetic, age: 45$\pm$15 years). PSL was automatically detected and it was higher for diabetic patients (p=0.01) at 4.26 $\pm$ 8.32 compared to 3.19 $\pm$ 3.62 for non-diabetic patients. The PancAP model achieved the highest Dice score of 0.79 $\pm$ 0.17 and lowest ASSD error of 1.94 $\pm$ 2.63 mm (p$<$0.05). For predicting T2DM, a multivariate model trained with CT biomarkers attained 0.90 AUC, 66.7\% sensitivity, and 91.9\% specificity. Our results suggest that PSL is useful for T2DM screening and could potentially help predict the early onset of T2DM.
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
Deep Learning Segmentation of Ascites on Abdominal CT Scans for Automatic Volume Quantification
Jun 23, 2024



Abstract:Purpose: To evaluate the performance of an automated deep learning method in detecting ascites and subsequently quantifying its volume in patients with liver cirrhosis and ovarian cancer. Materials and Methods: This retrospective study included contrast-enhanced and non-contrast abdominal-pelvic CT scans of patients with cirrhotic ascites and patients with ovarian cancer from two institutions, National Institutes of Health (NIH) and University of Wisconsin (UofW). The model, trained on The Cancer Genome Atlas Ovarian Cancer dataset (mean age, 60 years +/- 11 [s.d.]; 143 female), was tested on two internal (NIH-LC and NIH-OV) and one external dataset (UofW-LC). Its performance was measured by the Dice coefficient, standard deviations, and 95% confidence intervals, focusing on ascites volume in the peritoneal cavity. Results: On NIH-LC (25 patients; mean age, 59 years +/- 14 [s.d.]; 14 male) and NIH-OV (166 patients; mean age, 65 years +/- 9 [s.d.]; all female), the model achieved Dice scores of 0.855 +/- 0.061 (CI: 0.831-0.878) and 0.826 +/- 0.153 (CI: 0.764-0.887), with median volume estimation errors of 19.6% (IQR: 13.2-29.0) and 5.3% (IQR: 2.4-9.7) respectively. On UofW-LC (124 patients; mean age, 46 years +/- 12 [s.d.]; 73 female), the model had a Dice score of 0.830 +/- 0.107 (CI: 0.798-0.863) and median volume estimation error of 9.7% (IQR: 4.5-15.1). The model showed strong agreement with expert assessments, with r^2 values of 0.79, 0.98, and 0.97 across the test sets. Conclusion: The proposed deep learning method performed well in segmenting and quantifying the volume of ascites in concordance with expert radiologist assessments.
Lesion classification by model-based feature extraction: A differential affine invariant model of soft tissue elasticity
May 27, 2022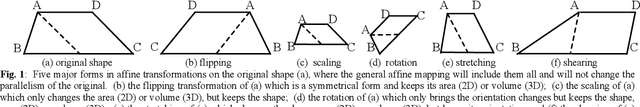
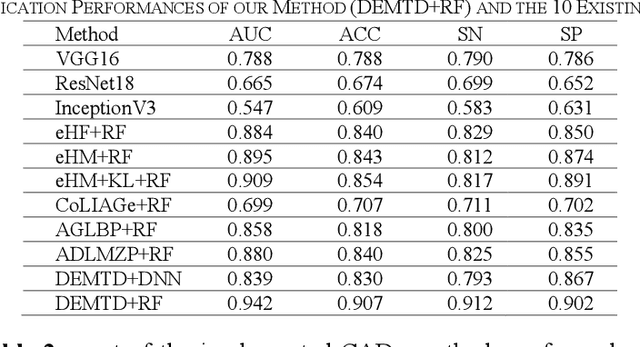
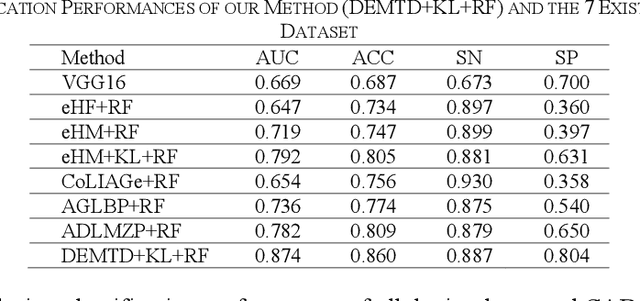
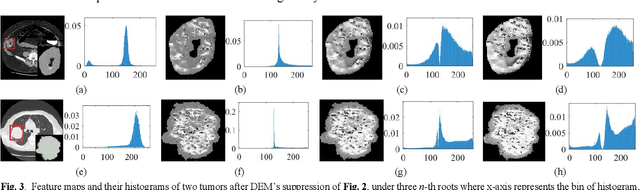
Abstract:The elasticity of soft tissues has been widely considered as a characteristic property to differentiate between healthy and vicious tissues and, therefore, motivated several elasticity imaging modalities, such as Ultrasound Elastography, Magnetic Resonance Elastography, and Optical Coherence Elastography. This paper proposes an alternative approach of modeling the elasticity using Computed Tomography (CT) imaging modality for model-based feature extraction machine learning (ML) differentiation of lesions. The model describes a dynamic non-rigid (or elastic) deformation in differential manifold to mimic the soft tissues elasticity under wave fluctuation in vivo. Based on the model, three local deformation invariants are constructed by two tensors defined by the first and second order derivatives from the CT images and used to generate elastic feature maps after normalization via a novel signal suppression method. The model-based elastic image features are extracted from the feature maps and fed to machine learning to perform lesion classifications. Two pathologically proven image datasets of colon polyps (44 malignant and 43 benign) and lung nodules (46 malignant and 20 benign) were used to evaluate the proposed model-based lesion classification. The outcomes of this modeling approach reached the score of area under the curve of the receiver operating characteristics of 94.2 % for the polyps and 87.4 % for the nodules, resulting in an average gain of 5 % to 30 % over ten existing state-of-the-art lesion classification methods. The gains by modeling tissue elasticity for ML differentiation of lesions are striking, indicating the great potential of exploring the modeling strategy to other tissue properties for ML differentiation of lesions.
Cross-Domain Medical Image Translation by Shared Latent Gaussian Mixture Model
Jul 14, 2020



Abstract:Current deep learning based segmentation models often generalize poorly between domains due to insufficient training data. In real-world clinical applications, cross-domain image analysis tools are in high demand since medical images from different domains are often needed to achieve a precise diagnosis. An important example in radiology is generalizing from non-contrast CT to contrast enhanced CTs. Contrast enhanced CT scans at different phases are used to enhance certain pathologies or organs. Many existing cross-domain image-to-image translation models have been shown to improve cross-domain segmentation of large organs. However, such models lack the ability to preserve fine structures during the translation process, which is significant for many clinical applications, such as segmenting small calcified plaques in the aorta and pelvic arteries. In order to preserve fine structures during medical image translation, we propose a patch-based model using shared latent variables from a Gaussian mixture model. We compare our image translation framework to several state-of-the-art methods on cross-domain image translation and show our model does a better job preserving fine structures. The superior performance of our model is verified by performing two tasks with the translated images - detection and segmentation of aortic plaques and pancreas segmentation. We expect the utility of our framework will extend to other problems beyond segmentation due to the improved quality of the generated images and enhanced ability to preserve small structures.
Image Translation by Latent Union of Subspaces for Cross-Domain Plaque Detection
May 22, 2020

Abstract:Calcified plaque in the aorta and pelvic arteries is associated with coronary artery calcification and is a strong predictor of heart attack. Current calcified plaque detection models show poor generalizability to different domains (ie. pre-contrast vs. post-contrast CT scans). Many recent works have shown how cross domain object detection can be improved using an image translation model which translates between domains using a single shared latent space. However, while current image translation models do a good job preserving global/intermediate level structures they often have trouble preserving tiny structures. In medical imaging applications, preserving small structures is important since these structures can carry information which is highly relevant for disease diagnosis. Recent works on image reconstruction show that complex real-world images are better reconstructed using a union of subspaces approach. Since small image patches are used to train the image translation model, it makes sense to enforce that each patch be represented by a linear combination of subspaces which may correspond to the different parts of the body present in that patch. Motivated by this, we propose an image translation network using a shared union of subspaces constraint and show our approach preserves subtle structures (plaques) better than the conventional method. We further applied our method to a cross domain plaque detection task and show significant improvement compared to the state-of-the art method.
Accurately identifying vertebral levels in large datasets
Jan 28, 2020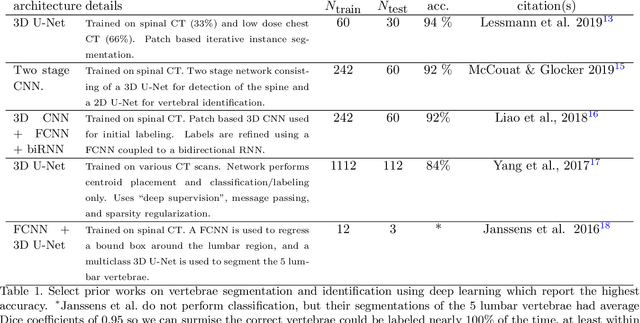
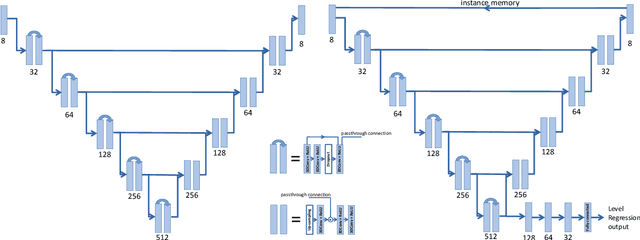
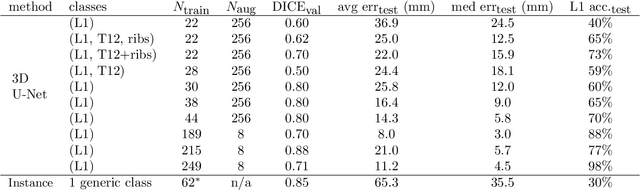
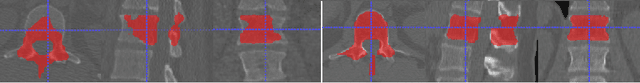
Abstract:The vertebral levels of the spine provide a useful coordinate system when making measurements of plaque, muscle, fat, and bone mineral density. Correctly classifying vertebral levels with high accuracy is challenging due to the similar appearance of each vertebra, the curvature of the spine, and the possibility of anomalies such as fractured vertebrae, implants, lumbarization of the sacrum, and sacralization of L5. The goal of this work is to develop a system that can accurately and robustly identify the L1 level in large heterogeneous datasets. The first approach we study is using a 3D U-Net to segment the L1 vertebra directly using the entire scan volume to provide context. We also tested models for two class segmentation of L1 and T12 and a three class segmentation of L1, T12 and the rib attached to T12. By increasing the number of training examples to 249 scans using pseudo-segmentations from an in-house segmentation tool we were able to achieve 98% accuracy with respect to identifying the L1 vertebra, with an average error of 4.5 mm in the craniocaudal level. We next developed an algorithm which performs iterative instance segmentation and classification of the entire spine with a 3D U-Net. We found the instance based approach was able to yield better segmentations of nearly the entire spine, but had lower classification accuracy for L1.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge