Veit Sandfort
Accurately identifying vertebral levels in large datasets
Jan 28, 2020
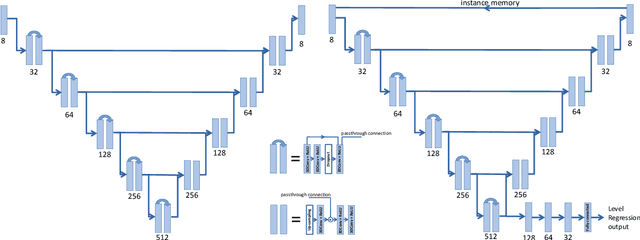
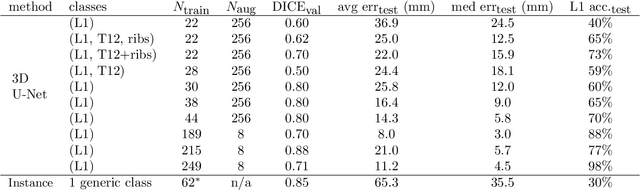
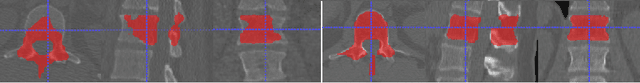
Abstract:The vertebral levels of the spine provide a useful coordinate system when making measurements of plaque, muscle, fat, and bone mineral density. Correctly classifying vertebral levels with high accuracy is challenging due to the similar appearance of each vertebra, the curvature of the spine, and the possibility of anomalies such as fractured vertebrae, implants, lumbarization of the sacrum, and sacralization of L5. The goal of this work is to develop a system that can accurately and robustly identify the L1 level in large heterogeneous datasets. The first approach we study is using a 3D U-Net to segment the L1 vertebra directly using the entire scan volume to provide context. We also tested models for two class segmentation of L1 and T12 and a three class segmentation of L1, T12 and the rib attached to T12. By increasing the number of training examples to 249 scans using pseudo-segmentations from an in-house segmentation tool we were able to achieve 98% accuracy with respect to identifying the L1 vertebra, with an average error of 4.5 mm in the craniocaudal level. We next developed an algorithm which performs iterative instance segmentation and classification of the entire spine with a 3D U-Net. We found the instance based approach was able to yield better segmentations of nearly the entire spine, but had lower classification accuracy for L1.
TUNA-Net: Task-oriented UNsupervised Adversarial Network for Disease Recognition in Cross-Domain Chest X-rays
Aug 21, 2019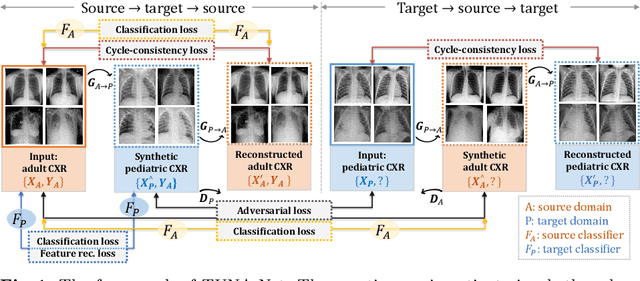
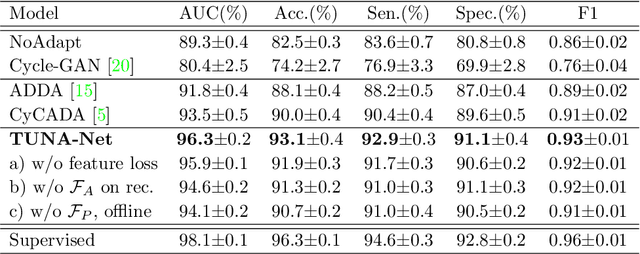
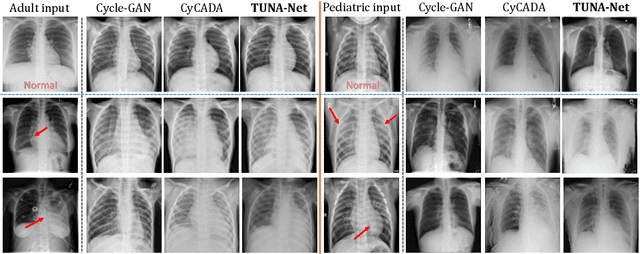
Abstract:In this work, we exploit the unsupervised domain adaptation problem for radiology image interpretation across domains. Specifically, we study how to adapt the disease recognition model from a labeled source domain to an unlabeled target domain, so as to reduce the effort of labeling each new dataset. To address the shortcoming of cross-domain, unpaired image-to-image translation methods which typically ignore class-specific semantics, we propose a task-driven, discriminatively trained, cycle-consistent generative adversarial network, termed TUNA-Net. It is able to preserve 1) low-level details, 2) high-level semantic information and 3) mid-level feature representation during the image-to-image translation process, to favor the target disease recognition task. The TUNA-Net framework is general and can be readily adapted to other learning tasks. We evaluate the proposed framework on two public chest X-ray datasets for pneumonia recognition. The TUNA-Net model can adapt labeled adult chest X-rays in the source domain such that they appear as if they were drawn from pediatric X-rays in the unlabeled target domain, while preserving the disease semantics. Extensive experiments show the superiority of the proposed method as compared to state-of-the-art unsupervised domain adaptation approaches. Notably, TUNA-Net achieves an AUC of 96.3% for pediatric pneumonia classification, which is very close to that of the supervised approach (98.1%), but without the need for labels on the target domain.
MULAN: Multitask Universal Lesion Analysis Network for Joint Lesion Detection, Tagging, and Segmentation
Aug 12, 2019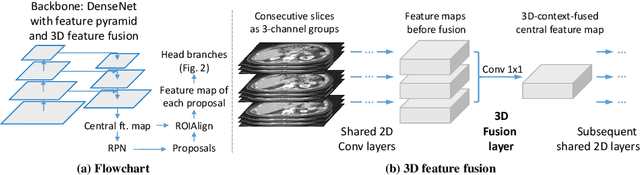
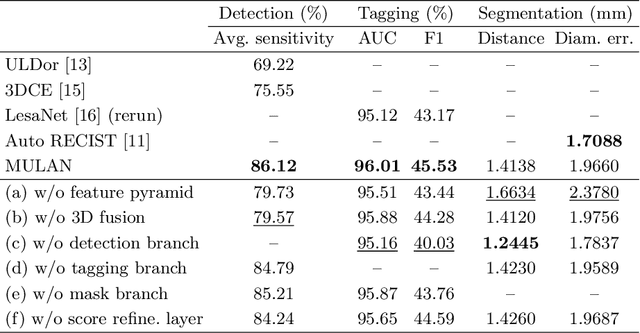
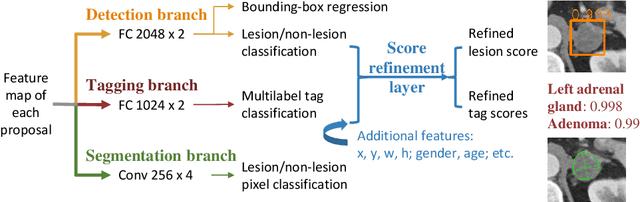
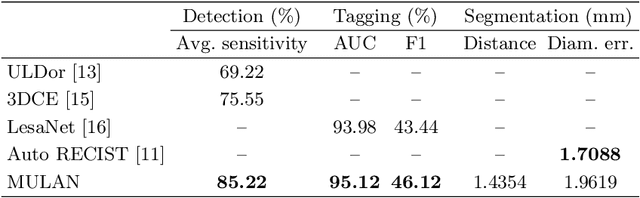
Abstract:When reading medical images such as a computed tomography (CT) scan, radiologists generally search across the image to find lesions, characterize and measure them, and then describe them in the radiological report. To automate this process, we propose a multitask universal lesion analysis network (MULAN) for joint detection, tagging, and segmentation of lesions in a variety of body parts, which greatly extends existing work of single-task lesion analysis on specific body parts. MULAN is based on an improved Mask R-CNN framework with three head branches and a 3D feature fusion strategy. It achieves the state-of-the-art accuracy in the detection and tagging tasks on the DeepLesion dataset, which contains 32K lesions in the whole body. We also analyze the relationship between the three tasks and show that tag predictions can improve detection accuracy via a score refinement layer.
A self-attention based deep learning method for lesion attribute detection from CT reports
Apr 30, 2019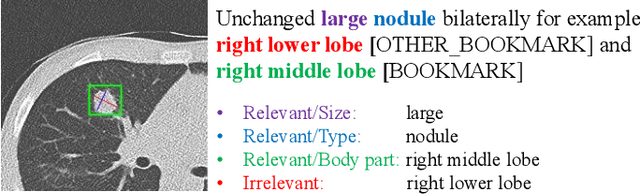
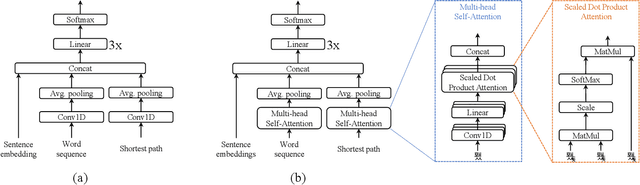
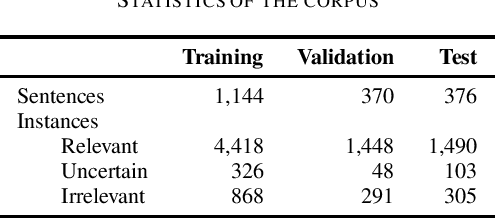
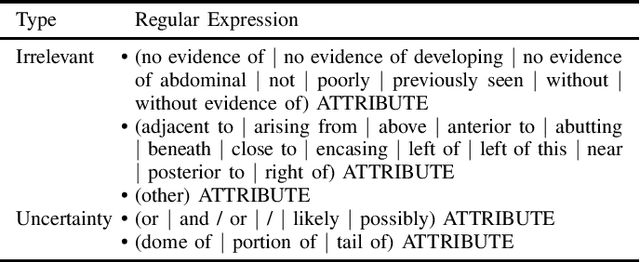
Abstract:In radiology, radiologists not only detect lesions from the medical image, but also describe them with various attributes such as their type, location, size, shape, and intensity. While these lesion attributes are rich and useful in many downstream clinical applications, how to extract them from the radiology reports is less studied. This paper outlines a novel deep learning method to automatically extract attributes of lesions of interest from the clinical text. Different from classical CNN models, we integrated the multi-head self-attention mechanism to handle the long-distance information in the sentence, and to jointly correlate different portions of sentence representation subspaces in parallel. Evaluation on an in-house corpus demonstrates that our method can achieve high performance with 0.848 in precision, 0.788 in recall, and 0.815 in F-score. The new method and constructed corpus will enable us to build automatic systems with a higher-level understanding of the radiological world.
Holistic and Comprehensive Annotation of Clinically Significant Findings on Diverse CT Images: Learning from Radiology Reports and Label Ontology
Apr 27, 2019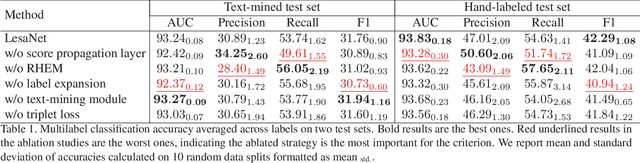
Abstract:In radiologists' routine work, one major task is to read a medical image, e.g., a CT scan, find significant lesions, and describe them in the radiology report. In this paper, we study the lesion description or annotation problem. Given a lesion image, our aim is to predict a comprehensive set of relevant labels, such as the lesion's body part, type, and attributes, which may assist downstream fine-grained diagnosis. To address this task, we first design a deep learning module to extract relevant semantic labels from the radiology reports associated with the lesion images. With the images and text-mined labels, we propose a lesion annotation network (LesaNet) based on a multilabel convolutional neural network (CNN) to learn all labels holistically. Hierarchical relations and mutually exclusive relations between the labels are leveraged to improve the label prediction accuracy. The relations are utilized in a label expansion strategy and a relational hard example mining algorithm. We also attach a simple score propagation layer on LesaNet to enhance recall and explore implicit relation between labels. Multilabel metric learning is combined with classification to enable interpretable prediction. We evaluated LesaNet on the public DeepLesion dataset, which contains over 32K diverse lesion images. Experiments show that LesaNet can precisely annotate the lesions using an ontology of 171 fine-grained labels with an average AUC of 0.9344.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge