Jiaxing Tan
Lesion classification by model-based feature extraction: A differential affine invariant model of soft tissue elasticity
May 27, 2022
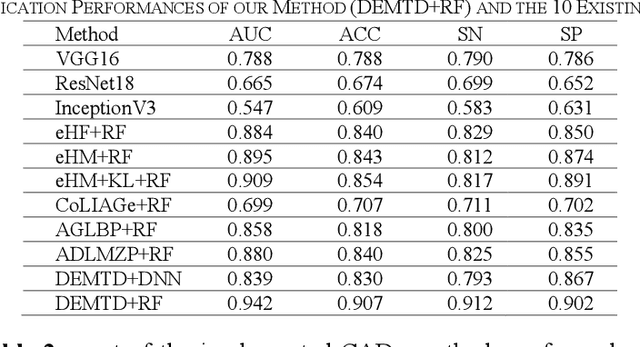
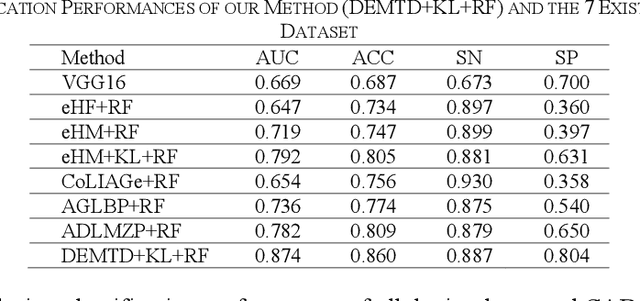
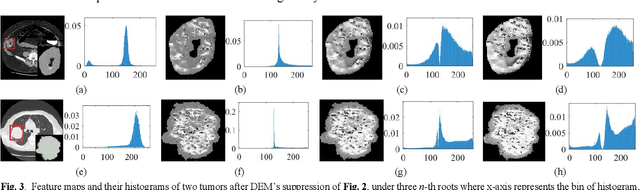
Abstract:The elasticity of soft tissues has been widely considered as a characteristic property to differentiate between healthy and vicious tissues and, therefore, motivated several elasticity imaging modalities, such as Ultrasound Elastography, Magnetic Resonance Elastography, and Optical Coherence Elastography. This paper proposes an alternative approach of modeling the elasticity using Computed Tomography (CT) imaging modality for model-based feature extraction machine learning (ML) differentiation of lesions. The model describes a dynamic non-rigid (or elastic) deformation in differential manifold to mimic the soft tissues elasticity under wave fluctuation in vivo. Based on the model, three local deformation invariants are constructed by two tensors defined by the first and second order derivatives from the CT images and used to generate elastic feature maps after normalization via a novel signal suppression method. The model-based elastic image features are extracted from the feature maps and fed to machine learning to perform lesion classifications. Two pathologically proven image datasets of colon polyps (44 malignant and 43 benign) and lung nodules (46 malignant and 20 benign) were used to evaluate the proposed model-based lesion classification. The outcomes of this modeling approach reached the score of area under the curve of the receiver operating characteristics of 94.2 % for the polyps and 87.4 % for the nodules, resulting in an average gain of 5 % to 30 % over ten existing state-of-the-art lesion classification methods. The gains by modeling tissue elasticity for ML differentiation of lesions are striking, indicating the great potential of exploring the modeling strategy to other tissue properties for ML differentiation of lesions.
Cross-modal Center Loss
Aug 08, 2020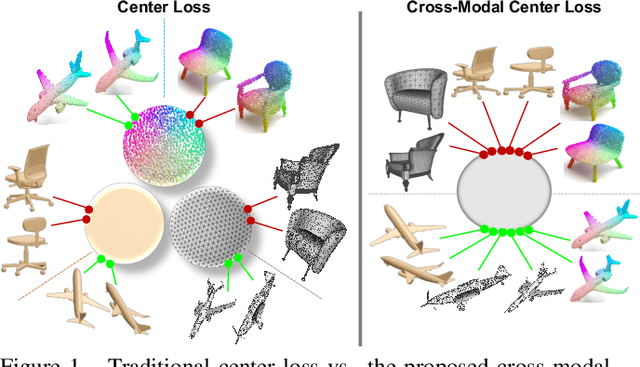
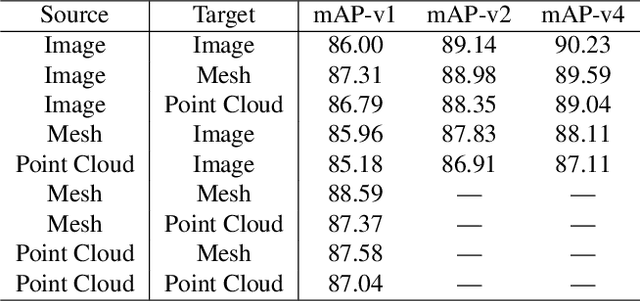
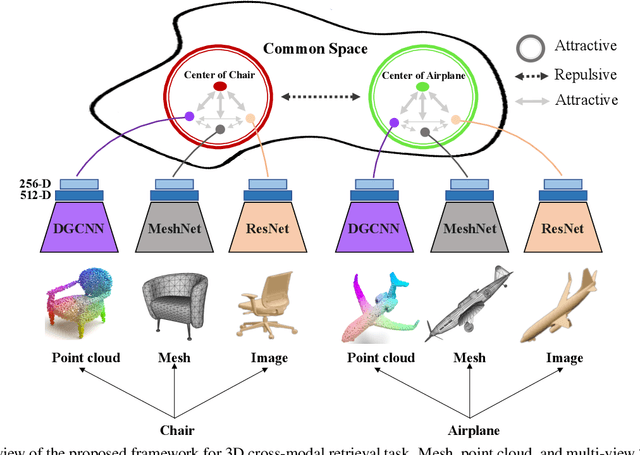
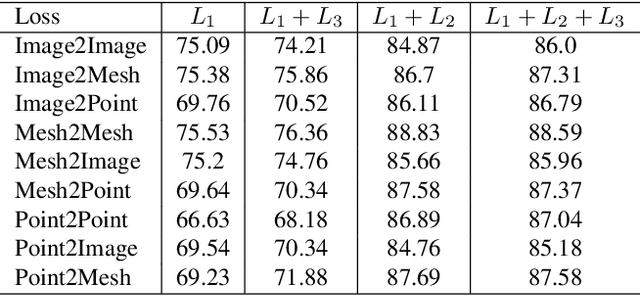
Abstract:Cross-modal retrieval aims to learn discriminative and modal-invariant features for data from different modalities. Unlike the existing methods which usually learn from the features extracted by offline networks, in this paper, we propose an approach to jointly train the components of cross-modal retrieval framework with metadata, and enable the network to find optimal features. The proposed end-to-end framework is updated with three loss functions: 1) a novel cross-modal center loss to eliminate cross-modal discrepancy, 2) cross-entropy loss to maximize inter-class variations, and 3) mean-square-error loss to reduce modality variations. In particular, our proposed cross-modal center loss minimizes the distances of features from objects belonging to the same class across all modalities. Extensive experiments have been conducted on the retrieval tasks across multi-modalities, including 2D image, 3D point cloud, and mesh data. The proposed framework significantly outperforms the state-of-the-art methods on the ModelNet40 dataset.
LGAN: Lung Segmentation in CT Scans Using Generative Adversarial Network
Jan 11, 2019



Abstract:Lung segmentation in computerized tomography (CT) images is an important procedure in various lung disease diagnosis. Most of the current lung segmentation approaches are performed through a series of procedures with manually empirical parameter adjustments in each step. Pursuing an automatic segmentation method with fewer steps, in this paper, we propose a novel deep learning Generative Adversarial Network (GAN) based lung segmentation schema, which we denote as LGAN. Our proposed schema can be generalized to different kinds of neural networks for lung segmentation in CT images and is evaluated on a dataset containing 220 individual CT scans with two metrics: segmentation quality and shape similarity. Also, we compared our work with current state of the art methods. The results obtained with this study demonstrate that the proposed LGAN schema can be used as a promising tool for automatic lung segmentation due to its simplified procedure as well as its good performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge