Weiwei Cao
Large-scale and Fine-grained Vision-language Pre-training for Enhanced CT Image Understanding
Jan 24, 2025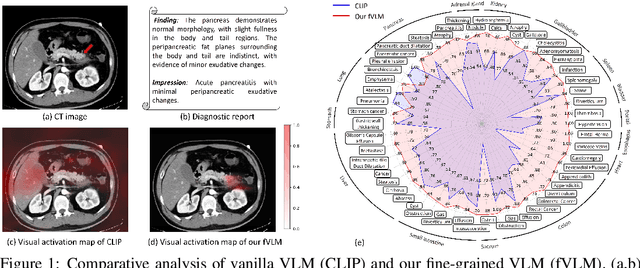
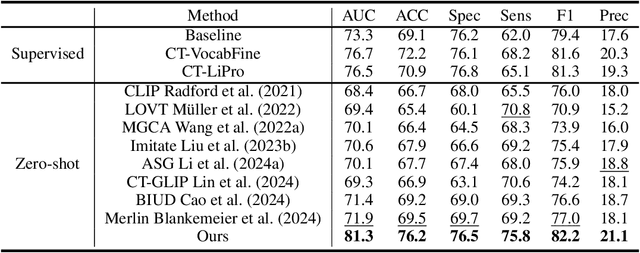
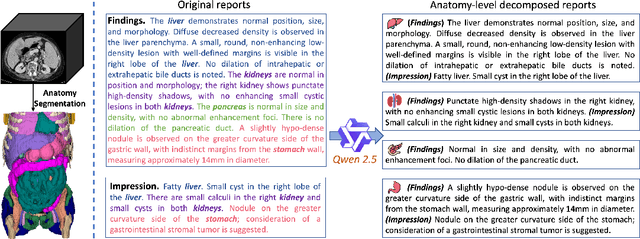
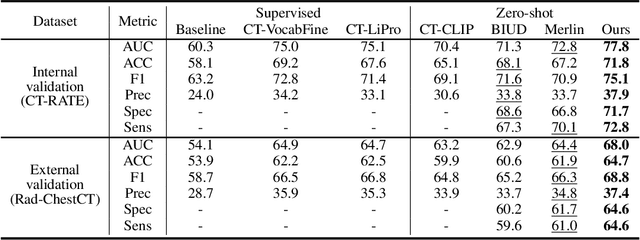
Abstract:Artificial intelligence (AI) shows great potential in assisting radiologists to improve the efficiency and accuracy of medical image interpretation and diagnosis. However, a versatile AI model requires large-scale data and comprehensive annotations, which are often impractical in medical settings. Recent studies leverage radiology reports as a naturally high-quality supervision for medical images, using contrastive language-image pre-training (CLIP) to develop language-informed models for radiological image interpretation. Nonetheless, these approaches typically contrast entire images with reports, neglecting the local associations between imaging regions and report sentences, which may undermine model performance and interoperability. In this paper, we propose a fine-grained vision-language model (fVLM) for anatomy-level CT image interpretation. Specifically, we explicitly match anatomical regions of CT images with corresponding descriptions in radiology reports and perform contrastive pre-training for each anatomy individually. Fine-grained alignment, however, faces considerable false-negative challenges, mainly from the abundance of anatomy-level healthy samples and similarly diseased abnormalities. To tackle this issue, we propose identifying false negatives of both normal and abnormal samples and calibrating contrastive learning from patient-level to disease-aware pairing. We curated the largest CT dataset to date, comprising imaging and report data from 69,086 patients, and conducted a comprehensive evaluation of 54 major and important disease diagnosis tasks across 15 main anatomies. Experimental results demonstrate the substantial potential of fVLM in versatile medical image interpretation. In the zero-shot classification task, we achieved an average AUC of 81.3% on 54 diagnosis tasks, surpassing CLIP and supervised methods by 12.9% and 8.0%, respectively.
Bootstrapping Chest CT Image Understanding by Distilling Knowledge from X-ray Expert Models
Apr 07, 2024



Abstract:Radiologists highly desire fully automated versatile AI for medical imaging interpretation. However, the lack of extensively annotated large-scale multi-disease datasets has hindered the achievement of this goal. In this paper, we explore the feasibility of leveraging language as a naturally high-quality supervision for chest CT imaging. In light of the limited availability of image-report pairs, we bootstrap the understanding of 3D chest CT images by distilling chest-related diagnostic knowledge from an extensively pre-trained 2D X-ray expert model. Specifically, we propose a language-guided retrieval method to match each 3D CT image with its semantically closest 2D X-ray image, and perform pair-wise and semantic relation knowledge distillation. Subsequently, we use contrastive learning to align images and reports within the same patient while distinguishing them from the other patients. However, the challenge arises when patients have similar semantic diagnoses, such as healthy patients, potentially confusing if treated as negatives. We introduce a robust contrastive learning that identifies and corrects these false negatives. We train our model with over 12,000 pairs of chest CT images and radiology reports. Extensive experiments across multiple scenarios, including zero-shot learning, report generation, and fine-tuning processes, demonstrate the model's feasibility in interpreting chest CT images.
Using Large Context for Kidney Multi-Structure Segmentation from CTA Images
Aug 10, 2022


Abstract:Accurate and automated segmentation of multi-structure (i.e., kidneys, renal tu-mors, arteries, and veins) from 3D CTA is one of the most important tasks for surgery-based renal cancer treatment (e.g., laparoscopic partial nephrectomy). This paper briefly presents the main technique details of the multi-structure seg-mentation method in MICCAI 2022 KIPA challenge. The main contribution of this paper is that we design the 3D UNet with the large context information cap-turing capability. Our method ranked eighth on the MICCAI 2022 KIPA chal-lenge open testing dataset with a mean position of 8.2. Our code and trained models are publicly available at https://github.com/fengjiejiejiejie/kipa22_nnunet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge