Tingbo Liang
Large-scale and Fine-grained Vision-language Pre-training for Enhanced CT Image Understanding
Jan 24, 2025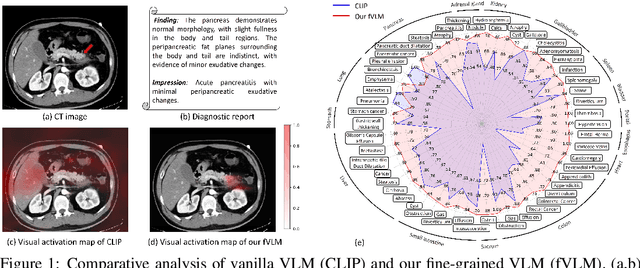
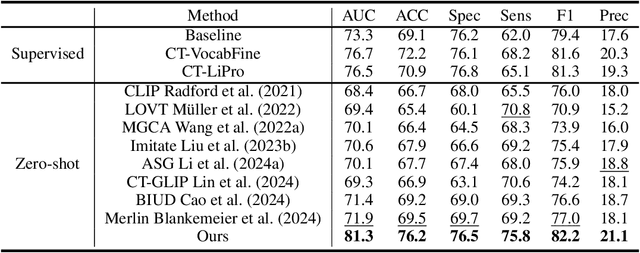
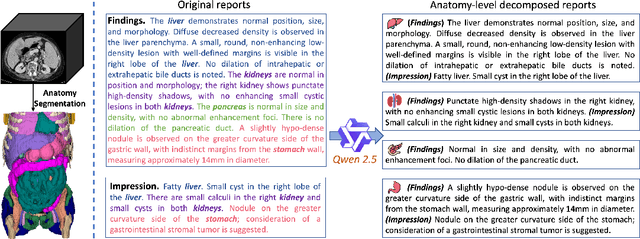
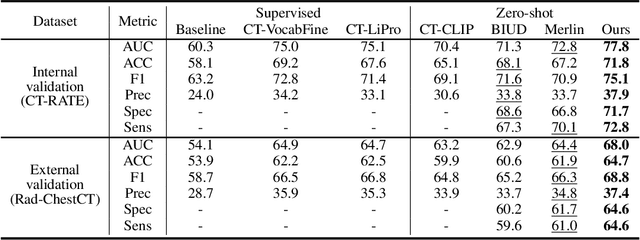
Abstract:Artificial intelligence (AI) shows great potential in assisting radiologists to improve the efficiency and accuracy of medical image interpretation and diagnosis. However, a versatile AI model requires large-scale data and comprehensive annotations, which are often impractical in medical settings. Recent studies leverage radiology reports as a naturally high-quality supervision for medical images, using contrastive language-image pre-training (CLIP) to develop language-informed models for radiological image interpretation. Nonetheless, these approaches typically contrast entire images with reports, neglecting the local associations between imaging regions and report sentences, which may undermine model performance and interoperability. In this paper, we propose a fine-grained vision-language model (fVLM) for anatomy-level CT image interpretation. Specifically, we explicitly match anatomical regions of CT images with corresponding descriptions in radiology reports and perform contrastive pre-training for each anatomy individually. Fine-grained alignment, however, faces considerable false-negative challenges, mainly from the abundance of anatomy-level healthy samples and similarly diseased abnormalities. To tackle this issue, we propose identifying false negatives of both normal and abnormal samples and calibrating contrastive learning from patient-level to disease-aware pairing. We curated the largest CT dataset to date, comprising imaging and report data from 69,086 patients, and conducted a comprehensive evaluation of 54 major and important disease diagnosis tasks across 15 main anatomies. Experimental results demonstrate the substantial potential of fVLM in versatile medical image interpretation. In the zero-shot classification task, we achieved an average AUC of 81.3% on 54 diagnosis tasks, surpassing CLIP and supervised methods by 12.9% and 8.0%, respectively.
Deep learning to estimate the physical proportion of infected region of lung for COVID-19 pneumonia with CT image set
Jun 09, 2020



Abstract:Utilizing computed tomography (CT) images to quickly estimate the severity of cases with COVID-19 is one of the most straightforward and efficacious methods. Two tasks were studied in this present paper. One was to segment the mask of intact lung in case of pneumonia. Another was to generate the masks of regions infected by COVID-19. The masks of these two parts of images then were converted to corresponding volumes to calculate the physical proportion of infected region of lung. A total of 129 CT image set were herein collected and studied. The intrinsic Hounsfiled value of CT images was firstly utilized to generate the initial dirty version of labeled masks both for intact lung and infected regions. Then, the samples were carefully adjusted and improved by two professional radiologists to generate the final training set and test benchmark. Two deep learning models were evaluated: UNet and 2.5D UNet. For the segment of infected regions, a deep learning based classifier was followed to remove unrelated blur-edged regions that were wrongly segmented out such as air tube and blood vessel tissue etc. For the segmented masks of intact lung and infected regions, the best method could achieve 0.972 and 0.757 measure in mean Dice similarity coefficient on our test benchmark. As the overall proportion of infected region of lung, the final result showed 0.961 (Pearson's correlation coefficient) and 11.7% (mean absolute percent error). The instant proportion of infected regions of lung could be used as a visual evidence to assist clinical physician to determine the severity of the case. Furthermore, a quantified report of infected regions can help predict the prognosis for COVID-19 cases which were scanned periodically within the treatment cycle.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge