Mingxin Zheng
VILA-M3: Enhancing Vision-Language Models with Medical Expert Knowledge
Nov 19, 2024



Abstract:Generalist vision language models (VLMs) have made significant strides in computer vision, but they fall short in specialized fields like healthcare, where expert knowledge is essential. In traditional computer vision tasks, creative or approximate answers may be acceptable, but in healthcare, precision is paramount.Current large multimodal models like Gemini and GPT-4o are insufficient for medical tasks due to their reliance on memorized internet knowledge rather than the nuanced expertise required in healthcare. VLMs are usually trained in three stages: vision pre-training, vision-language pre-training, and instruction fine-tuning (IFT). IFT has been typically applied using a mixture of generic and healthcare data. In contrast, we propose that for medical VLMs, a fourth stage of specialized IFT is necessary, which focuses on medical data and includes information from domain expert models. Domain expert models developed for medical use are crucial because they are specifically trained for certain clinical tasks, e.g. to detect tumors and classify abnormalities through segmentation and classification, which learn fine-grained features of medical data$-$features that are often too intricate for a VLM to capture effectively especially in radiology. This paper introduces a new framework, VILA-M3, for medical VLMs that utilizes domain knowledge via expert models. Through our experiments, we show an improved state-of-the-art (SOTA) performance with an average improvement of ~9% over the prior SOTA model Med-Gemini and ~6% over models trained on the specific tasks. Our approach emphasizes the importance of domain expertise in creating precise, reliable VLMs for medical applications.
MONAI: An open-source framework for deep learning in healthcare
Nov 04, 2022



Abstract:Artificial Intelligence (AI) is having a tremendous impact across most areas of science. Applications of AI in healthcare have the potential to improve our ability to detect, diagnose, prognose, and intervene on human disease. For AI models to be used clinically, they need to be made safe, reproducible and robust, and the underlying software framework must be aware of the particularities (e.g. geometry, physiology, physics) of medical data being processed. This work introduces MONAI, a freely available, community-supported, and consortium-led PyTorch-based framework for deep learning in healthcare. MONAI extends PyTorch to support medical data, with a particular focus on imaging, and provide purpose-specific AI model architectures, transformations and utilities that streamline the development and deployment of medical AI models. MONAI follows best practices for software-development, providing an easy-to-use, robust, well-documented, and well-tested software framework. MONAI preserves the simple, additive, and compositional approach of its underlying PyTorch libraries. MONAI is being used by and receiving contributions from research, clinical and industrial teams from around the world, who are pursuing applications spanning nearly every aspect of healthcare.
Automatic ultrasound vessel segmentation with deep spatiotemporal context learning
Nov 03, 2021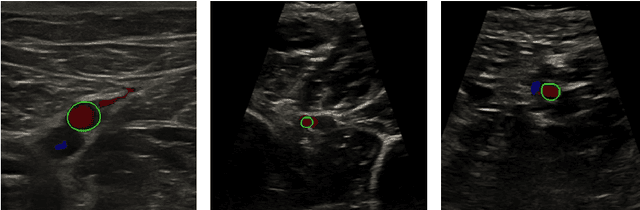
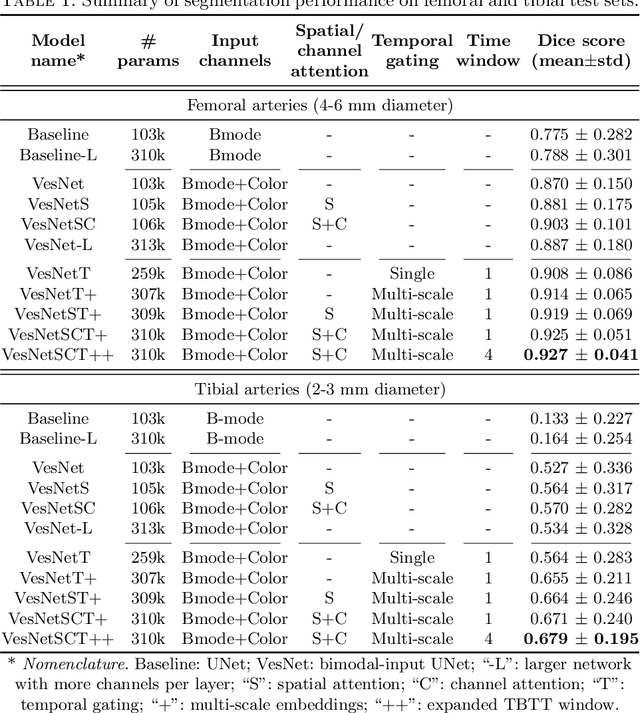
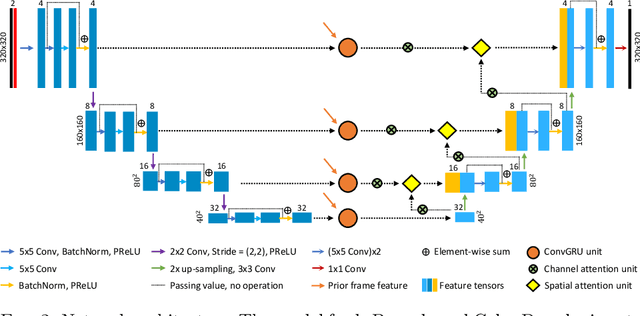
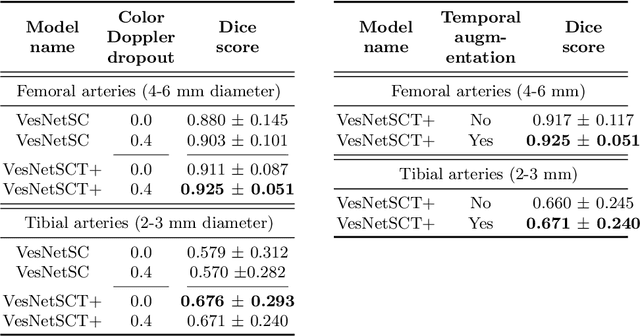
Abstract:Accurate, real-time segmentation of vessel structures in ultrasound image sequences can aid in the measurement of lumen diameters and assessment of vascular diseases. This, however, remains a challenging task, particularly for extremely small vessels that are difficult to visualize. We propose to leverage the rich spatiotemporal context available in ultrasound to improve segmentation of small-scale lower-extremity arterial vasculature. We describe efficient deep learning methods that incorporate temporal, spatial, and feature-aware contextual embeddings at multiple resolution scales while jointly utilizing information from B-mode and Color Doppler signals. Evaluating on femoral and tibial artery scans performed on healthy subjects by an expert ultrasonographer, and comparing to consensus expert ground-truth annotations of inner lumen boundaries, we demonstrate real-time segmentation using the context-aware models and show that they significantly outperform comparable baseline approaches.
RiWalk: Fast Structural Node Embedding via Role Identification
Oct 15, 2019



Abstract:Nodes performing different functions in a network have different roles, and these roles can be gleaned from the structure of the network. Learning latent representations for the roles of nodes helps to understand the network and to transfer knowledge across networks. However, most existing structural embedding approaches suffer from high computation and space cost or rely on heuristic feature engineering. Here we propose RiWalk, a flexible paradigm for learning structural node representations. It decouples the structural embedding problem into a role identification procedure and a network embedding procedure. Through role identification, rooted kernels with structural dependencies kept are built to better integrate network embedding methods. To demonstrate the effectiveness of RiWalk, we develop two different role identification methods named RiWalk-SP and RiWalk-WL respectively and employ random walk based network embedding methods. Experiments on within-network classification tasks show that our proposed algorithms achieve comparable performance with other baselines while being an order of magnitude more efficient. Besides, we also conduct across-network role classification tasks. The results show potential of structural embeddings in transfer learning. RiWalk is also scalable, making it capable of capturing structural roles in massive networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge