Xueting Han
Beyond Two-Stage Training: Cooperative SFT and RL for LLM Reasoning
Sep 08, 2025Abstract:Reinforcement learning (RL) has proven effective in incentivizing the reasoning abilities of large language models (LLMs), but suffers from severe efficiency challenges due to its trial-and-error nature. While the common practice employs supervised fine-tuning (SFT) as a warm-up stage for RL, this decoupled two-stage approach limits interaction between SFT and RL, thereby constraining overall effectiveness. This study introduces a novel method for learning reasoning models that employs bilevel optimization to facilitate better cooperation between these training paradigms. By conditioning the SFT objective on the optimal RL policy, our approach enables SFT to meta-learn how to guide RL's optimization process. During training, the lower level performs RL updates while simultaneously receiving SFT supervision, and the upper level explicitly maximizes the cooperative gain-the performance advantage of joint SFT-RL training over RL alone. Empirical evaluations on five reasoning benchmarks demonstrate that our method consistently outperforms baselines and achieves a better balance between effectiveness and efficiency.
ReVeal: Self-Evolving Code Agents via Iterative Generation-Verification
Jun 13, 2025



Abstract:Recent advances in reinforcement learning (RL) with verifiable outcome rewards have significantly improved the reasoning capabilities of large language models (LLMs), especially when combined with multi-turn tool interactions. However, existing methods lack both meaningful verification signals from realistic environments and explicit optimization for verification, leading to unreliable self-verification. To address these limitations, we propose ReVeal, a multi-turn reinforcement learning framework that interleaves code generation with explicit self-verification and tool-based evaluation. ReVeal enables LLMs to autonomously generate test cases, invoke external tools for precise feedback, and improves performance via a customized RL algorithm with dense, per-turn rewards. As a result, ReVeal fosters the co-evolution of a model's generation and verification capabilities through RL training, expanding the reasoning boundaries of the base model, demonstrated by significant gains in Pass@k on LiveCodeBench. It also enables test-time scaling into deeper inference regimes, with code consistently evolving as the number of turns increases during inference, ultimately surpassing DeepSeek-R1-Zero-Qwen-32B. These findings highlight the promise of ReVeal as a scalable and effective paradigm for building more robust and autonomous AI agents.
Vulnerability-Aware Alignment: Mitigating Uneven Forgetting in Harmful Fine-Tuning
Jun 04, 2025Abstract:Harmful fine-tuning (HFT), performed directly on open-source LLMs or through Fine-tuning-as-a-Service, breaks safety alignment and poses significant threats. Existing methods aim to mitigate HFT risks by learning robust representation on alignment data or making harmful data unlearnable, but they treat each data sample equally, leaving data vulnerability patterns understudied. In this work, we reveal that certain subsets of alignment data are consistently more prone to forgetting during HFT across different fine-tuning tasks. Inspired by these findings, we propose Vulnerability-Aware Alignment (VAA), which estimates data vulnerability, partitions data into "vulnerable" and "invulnerable" groups, and encourages balanced learning using a group distributionally robust optimization (Group DRO) framework. Specifically, VAA learns an adversarial sampler that samples examples from the currently underperforming group and then applies group-dependent adversarial perturbations to the data during training, aiming to encourage a balanced learning process across groups. Experiments across four fine-tuning tasks demonstrate that VAA significantly reduces harmful scores while preserving downstream task performance, outperforming state-of-the-art baselines.
BSM: Small but Powerful Biological Sequence Model for Genes and Proteins
Oct 15, 2024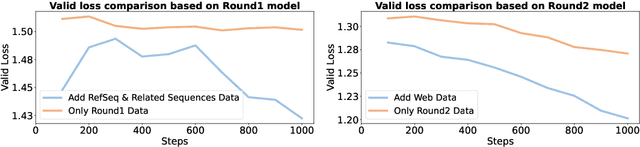

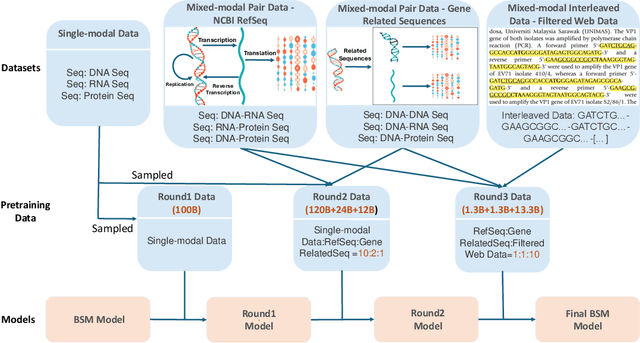
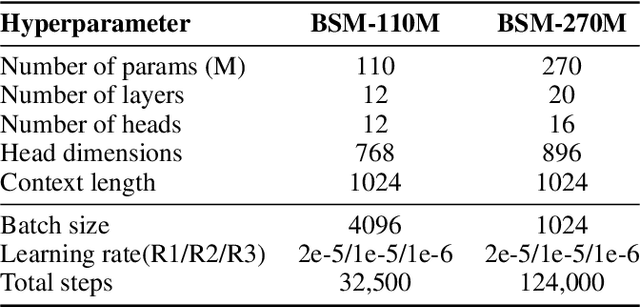
Abstract:Modeling biological sequences such as DNA, RNA, and proteins is crucial for understanding complex processes like gene regulation and protein synthesis. However, most current models either focus on a single type or treat multiple types of data separately, limiting their ability to capture cross-modal relationships. We propose that by learning the relationships between these modalities, the model can enhance its understanding of each type. To address this, we introduce BSM, a small but powerful mixed-modal biological sequence foundation model, trained on three types of data: RefSeq, Gene Related Sequences, and interleaved biological sequences from the web. These datasets capture the genetic flow, gene-protein relationships, and the natural co-occurrence of diverse biological data, respectively. By training on mixed-modal data, BSM significantly enhances learning efficiency and cross-modal representation, outperforming models trained solely on unimodal data. With only 110M parameters, BSM achieves performance comparable to much larger models across both single-modal and mixed-modal tasks, and uniquely demonstrates in-context learning capability for mixed-modal tasks, which is absent in existing models. Further scaling to 270M parameters demonstrates even greater performance gains, highlighting the potential of BSM as a significant advancement in multimodal biological sequence modeling.
CPL: Critical Planning Step Learning Boosts LLM Generalization in Reasoning Tasks
Sep 13, 2024



Abstract:Post-training large language models (LLMs) to develop reasoning capabilities has proven effective across diverse domains, such as mathematical reasoning and code generation. However, existing methods primarily focus on improving task-specific reasoning but have not adequately addressed the model's generalization capabilities across a broader range of reasoning tasks. To tackle this challenge, we introduce Critical Planning Step Learning (CPL), which leverages Monte Carlo Tree Search (MCTS) to explore diverse planning steps in multi-step reasoning tasks. Based on long-term outcomes, CPL learns step-level planning preferences to improve the model's planning capabilities and, consequently, its general reasoning capabilities. Furthermore, while effective in many scenarios for aligning LLMs, existing preference learning approaches like Direct Preference Optimization (DPO) struggle with complex multi-step reasoning tasks due to their inability to capture fine-grained supervision at each step. We propose Step-level Advantage Preference Optimization (Step-APO), which integrates an advantage estimate for step-level preference pairs obtained via MCTS into the DPO. This enables the model to more effectively learn critical intermediate planning steps, thereby further improving its generalization in reasoning tasks. Experimental results demonstrate that our method, trained exclusively on GSM8K and MATH, not only significantly improves performance on GSM8K (+10.5%) and MATH (+6.5%), but also enhances out-of-domain reasoning benchmarks, such as ARC-C (+4.0%), BBH (+1.8%), MMLU-STEM (+2.2%), and MMLU (+0.9%).
NutePrune: Efficient Progressive Pruning with Numerous Teachers for Large Language Models
Feb 15, 2024



Abstract:The considerable size of Large Language Models (LLMs) presents notable deployment challenges, particularly on resource-constrained hardware. Structured pruning, offers an effective means to compress LLMs, thereby reducing storage costs and enhancing inference speed for more efficient utilization. In this work, we study data-efficient and resource-efficient structure pruning methods to obtain smaller yet still powerful models. Knowledge Distillation is well-suited for pruning, as the intact model can serve as an excellent teacher for pruned students. However, it becomes challenging in the context of LLMs due to memory constraints. To address this, we propose an efficient progressive Numerous-teacher pruning method (NutePrune). NutePrune mitigates excessive memory costs by loading only one intact model and integrating it with various masks and LoRA modules, enabling it to seamlessly switch between teacher and student roles. This approach allows us to leverage numerous teachers with varying capacities to progressively guide the pruned model, enhancing overall performance. Extensive experiments across various tasks demonstrate the effectiveness of NutePrune. In LLaMA-7B zero-shot experiments, NutePrune retains 97.17% of the performance of the original model at 20% sparsity and 95.07% at 25% sparsity.
Parameter-efficient is not sufficient: Exploring Parameter, Memory, and Time Efficient Adapter Tuning for Dense Predictions
Jun 16, 2023Abstract:Pre-training & fine-tuning is a prevalent paradigm in computer vision (CV). Recently, parameter-efficient transfer learning (PETL) methods have shown promising performance in transferring knowledge from pre-trained models with only a few trainable parameters. Despite their success, the existing PETL methods in CV can be computationally expensive and require large amounts of memory and time cost during training, which limits low-resource users from conducting research and applications on large models. In this work, we propose Parameter, Memory, and Time Efficient Visual Adapter ($\mathrm{E^3VA}$) tuning to address this issue. We provide a gradient backpropagation highway for low-rank adapters which removes large gradient computations for the frozen pre-trained parameters, resulting in substantial savings of training memory and training time. Furthermore, we optimise the $\mathrm{E^3VA}$ structure for dense predictions tasks to promote model performance. Extensive experiments on COCO, ADE20K, and Pascal VOC benchmarks show that $\mathrm{E^3VA}$ can save up to 62.2% training memory and 26.2% training time on average, while achieving comparable performance to full fine-tuning and better performance than most PETL methods. Note that we can even train the Swin-Large-based Cascade Mask RCNN on GTX 1080Ti GPUs with less than 1.5% trainable parameters.
Time-aware Graph Structure Learning via Sequence Prediction on Temporal Graphs
Jun 13, 2023Abstract:Temporal Graph Learning, which aims to model the time-evolving nature of graphs, has gained increasing attention and achieved remarkable performance recently. However, in reality, graph structures are often incomplete and noisy, which hinders temporal graph networks (TGNs) from learning informative representations. Graph contrastive learning uses data augmentation to generate plausible variations of existing data and learn robust representations. However, rule-based augmentation approaches may be suboptimal as they lack learnability and fail to leverage rich information from downstream tasks. To address these issues, we propose a Time-aware Graph Structure Learning (TGSL) approach via sequence prediction on temporal graphs, which learns better graph structures for downstream tasks through adding potential temporal edges. In particular, it predicts time-aware context embedding based on previously observed interactions and uses the Gumble-Top-K to select the closest candidate edges to this context embedding. Additionally, several candidate sampling strategies are proposed to ensure both efficiency and diversity. Furthermore, we jointly learn the graph structure and TGNs in an end-to-end manner and perform inference on the refined graph. Extensive experiments on temporal link prediction benchmarks demonstrate that TGSL yields significant gains for the popular TGNs such as TGAT and GraphMixer, and it outperforms other contrastive learning methods on temporal graphs. We will release the code in the future.
AdapterGNN: Efficient Delta Tuning Improves Generalization Ability in Graph Neural Networks
Apr 19, 2023



Abstract:Fine-tuning pre-trained models has recently yielded remarkable performance gains in graph neural networks (GNNs). In addition to pre-training techniques, inspired by the latest work in the natural language fields, more recent work has shifted towards applying effective fine-tuning approaches, such as parameter-efficient tuning (delta tuning). However, given the substantial differences between GNNs and transformer-based models, applying such approaches directly to GNNs proved to be less effective. In this paper, we present a comprehensive comparison of delta tuning techniques for GNNs and propose a novel delta tuning method specifically designed for GNNs, called AdapterGNN. AdapterGNN preserves the knowledge of the large pre-trained model and leverages highly expressive adapters for GNNs, which can adapt to downstream tasks effectively with only a few parameters, while also improving the model's generalization ability on the downstream tasks. Extensive experiments show that AdapterGNN achieves higher evaluation performance (outperforming full fine-tuning by 1.4% and 5.5% in the chemistry and biology domains respectively, with only 5% of its parameters tuned) and lower generalization gaps compared to full fine-tuning. Moreover, we empirically show that a larger GNN model can have a worse generalization ability, which differs from the trend observed in large language models. We have also provided a theoretical justification for delta tuning can improve the generalization ability of GNNs by applying generalization bounds.
Adaptive Transfer Learning on Graph Neural Networks
Jul 20, 2021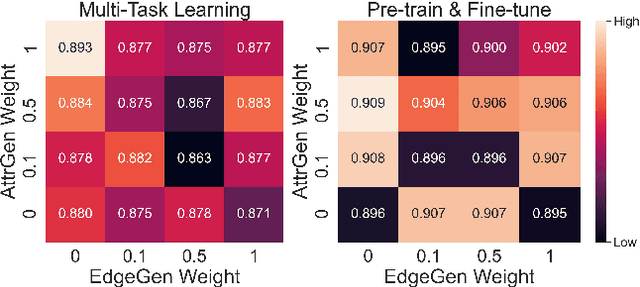
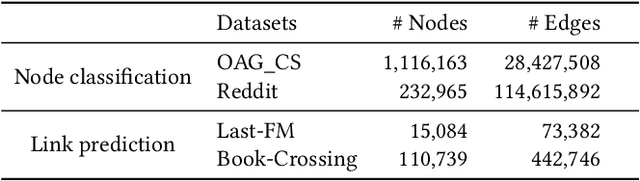


Abstract:Graph neural networks (GNNs) is widely used to learn a powerful representation of graph-structured data. Recent work demonstrates that transferring knowledge from self-supervised tasks to downstream tasks could further improve graph representation. However, there is an inherent gap between self-supervised tasks and downstream tasks in terms of optimization objective and training data. Conventional pre-training methods may be not effective enough on knowledge transfer since they do not make any adaptation for downstream tasks. To solve such problems, we propose a new transfer learning paradigm on GNNs which could effectively leverage self-supervised tasks as auxiliary tasks to help the target task. Our methods would adaptively select and combine different auxiliary tasks with the target task in the fine-tuning stage. We design an adaptive auxiliary loss weighting model to learn the weights of auxiliary tasks by quantifying the consistency between auxiliary tasks and the target task. In addition, we learn the weighting model through meta-learning. Our methods can be applied to various transfer learning approaches, it performs well not only in multi-task learning but also in pre-training and fine-tuning. Comprehensive experiments on multiple downstream tasks demonstrate that the proposed methods can effectively combine auxiliary tasks with the target task and significantly improve the performance compared to state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge