BSM: Small but Powerful Biological Sequence Model for Genes and Proteins
Paper and Code
Oct 15, 2024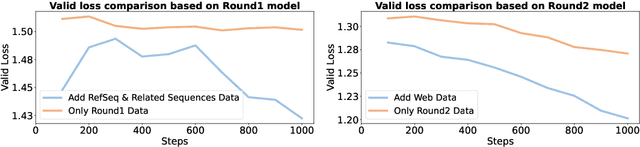

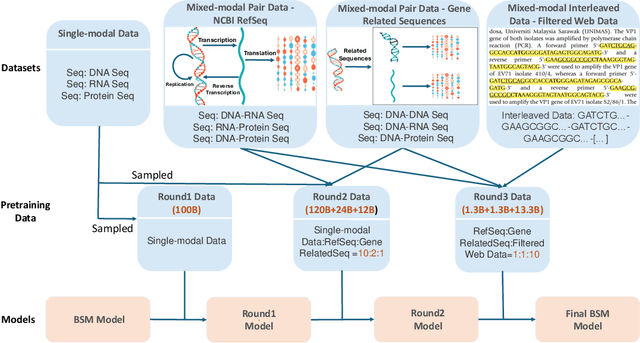
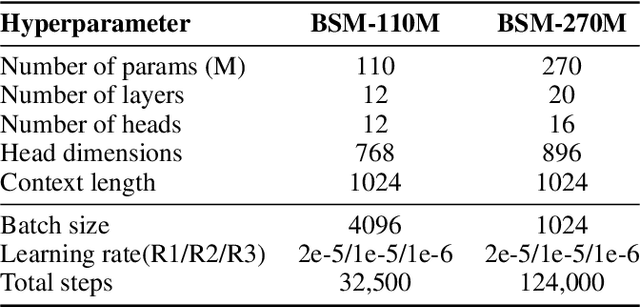
Modeling biological sequences such as DNA, RNA, and proteins is crucial for understanding complex processes like gene regulation and protein synthesis. However, most current models either focus on a single type or treat multiple types of data separately, limiting their ability to capture cross-modal relationships. We propose that by learning the relationships between these modalities, the model can enhance its understanding of each type. To address this, we introduce BSM, a small but powerful mixed-modal biological sequence foundation model, trained on three types of data: RefSeq, Gene Related Sequences, and interleaved biological sequences from the web. These datasets capture the genetic flow, gene-protein relationships, and the natural co-occurrence of diverse biological data, respectively. By training on mixed-modal data, BSM significantly enhances learning efficiency and cross-modal representation, outperforming models trained solely on unimodal data. With only 110M parameters, BSM achieves performance comparable to much larger models across both single-modal and mixed-modal tasks, and uniquely demonstrates in-context learning capability for mixed-modal tasks, which is absent in existing models. Further scaling to 270M parameters demonstrates even greater performance gains, highlighting the potential of BSM as a significant advancement in multimodal biological sequence modeling.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge