Min Zhu
MedKGI: Iterative Differential Diagnosis with Medical Knowledge Graphs and Information-Guided Inquiring
Dec 30, 2025Abstract:Recent advancements in Large Language Models (LLMs) have demonstrated significant promise in clinical diagnosis. However, current models struggle to emulate the iterative, diagnostic hypothesis-driven reasoning of real clinical scenarios. Specifically, current LLMs suffer from three critical limitations: (1) generating hallucinated medical content due to weak grounding in verified knowledge, (2) asking redundant or inefficient questions rather than discriminative ones that hinder diagnostic progress, and (3) losing coherence over multi-turn dialogues, leading to contradictory or inconsistent conclusions. To address these challenges, we propose MedKGI, a diagnostic framework grounded in clinical practices. MedKGI integrates a medical knowledge graph (KG) to constrain reasoning to validated medical ontologies, selects questions based on information gain to maximize diagnostic efficiency, and adopts an OSCE-format structured state to maintain consistent evidence tracking across turns. Experiments on clinical benchmarks show that MedKGI outperforms strong LLM baselines in both diagnostic accuracy and inquiry efficiency, improving dialogue efficiency by 30% on average while maintaining state-of-the-art accuracy.
PI-MFM: Physics-informed multimodal foundation model for solving partial differential equations
Dec 28, 2025Abstract:Partial differential equations (PDEs) govern a wide range of physical systems, and recent multimodal foundation models have shown promise for learning PDE solution operators across diverse equation families. However, existing multi-operator learning approaches are data-hungry and neglect physics during training. Here, we propose a physics-informed multimodal foundation model (PI-MFM) framework that directly enforces governing equations during pretraining and adaptation. PI-MFM takes symbolic representations of PDEs as the input, and automatically assembles PDE residual losses from the input expression via a vectorized derivative computation. These designs enable any PDE-encoding multimodal foundation model to be trained or adapted with unified physics-informed objectives across equation families. On a benchmark of 13 parametric one-dimensional time-dependent PDE families, PI-MFM consistently outperforms purely data-driven counterparts, especially with sparse labeled spatiotemporal points, partially observed time domains, or few labeled function pairs. Physics losses further improve robustness against noise, and simple strategies such as resampling collocation points substantially improve accuracy. We also analyze the accuracy, precision, and computational cost of automatic differentiation and finite differences for derivative computation within PI-MFM. Finally, we demonstrate zero-shot physics-informed fine-tuning to unseen PDE families: starting from a physics-informed pretrained model, adapting using only PDE residuals and initial/boundary conditions, without any labeled solution data, rapidly reduces test errors to around 1% and clearly outperforms physics-only training from scratch. These results show that PI-MFM provides a practical and scalable path toward data-efficient, transferable PDE solvers.
Towards Large-scale Generative Ranking
May 08, 2025
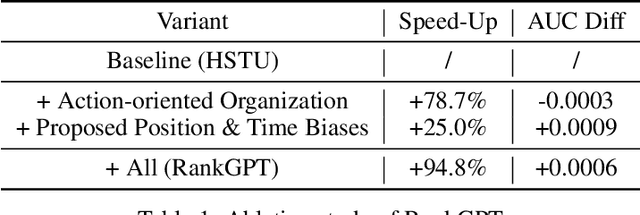
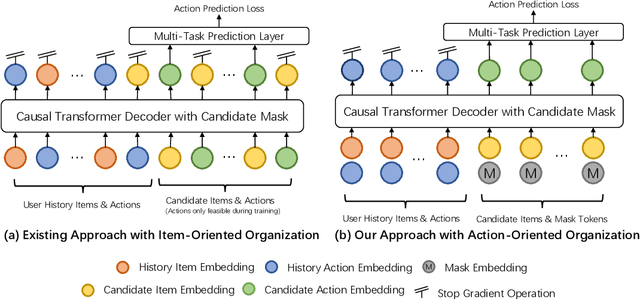

Abstract:Generative recommendation has recently emerged as a promising paradigm in information retrieval. However, generative ranking systems are still understudied, particularly with respect to their effectiveness and feasibility in large-scale industrial settings. This paper investigates this topic at the ranking stage of Xiaohongshu's Explore Feed, a recommender system that serves hundreds of millions of users. Specifically, we first examine how generative ranking outperforms current industrial recommenders. Through theoretical and empirical analyses, we find that the primary improvement in effectiveness stems from the generative architecture, rather than the training paradigm. To facilitate efficient deployment of generative ranking, we introduce GenRank, a novel generative architecture for ranking. We validate the effectiveness and efficiency of our solution through online A/B experiments. The results show that GenRank achieves significant improvements in user satisfaction with nearly equivalent computational resources compared to the existing production system.
COAST: Intelligent Time-Adaptive Neural Operators
Feb 12, 2025
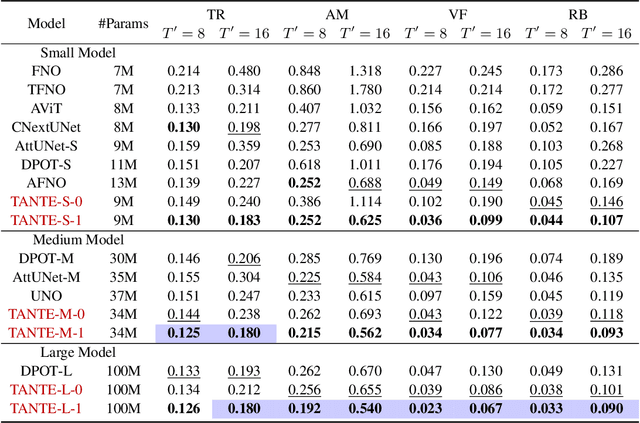
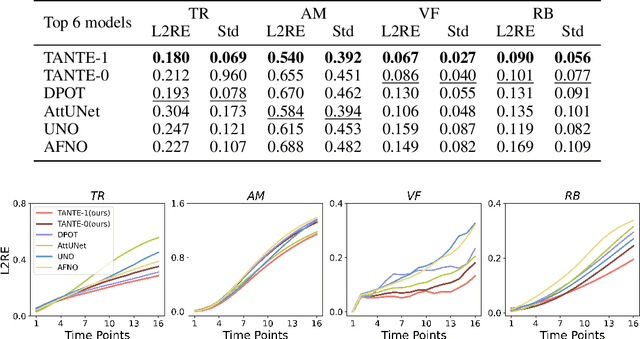
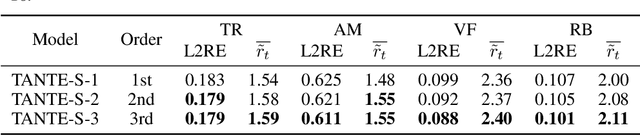
Abstract:We introduce Causal Operator with Adaptive Solver Transformer (COAST), a novel neural operator learning method that leverages a causal language model (CLM) framework to dynamically adapt time steps. Our method predicts both the evolution of a system and its optimal time step, intelligently balancing computational efficiency and accuracy. We find that COAST generates variable step sizes that correlate with the underlying system intrinsicities, both within and across dynamical systems. Within a single trajectory, smaller steps are taken in regions of high complexity, while larger steps are employed in simpler regions. Across different systems, more complex dynamics receive more granular time steps. Benchmarked on diverse systems with varied dynamics, COAST consistently outperforms state-of-the-art methods, achieving superior performance in both efficiency and accuracy. This work underscores the potential of CLM-based intelligent adaptive solvers for scalable operator learning of dynamical systems.
Interactive Medical Image Segmentation: A Benchmark Dataset and Baseline
Nov 19, 2024Abstract:Interactive Medical Image Segmentation (IMIS) has long been constrained by the limited availability of large-scale, diverse, and densely annotated datasets, which hinders model generalization and consistent evaluation across different models. In this paper, we introduce the IMed-361M benchmark dataset, a significant advancement in general IMIS research. First, we collect and standardize over 6.4 million medical images and their corresponding ground truth masks from multiple data sources. Then, leveraging the strong object recognition capabilities of a vision foundational model, we automatically generated dense interactive masks for each image and ensured their quality through rigorous quality control and granularity management. Unlike previous datasets, which are limited by specific modalities or sparse annotations, IMed-361M spans 14 modalities and 204 segmentation targets, totaling 361 million masks-an average of 56 masks per image. Finally, we developed an IMIS baseline network on this dataset that supports high-quality mask generation through interactive inputs, including clicks, bounding boxes, text prompts, and their combinations. We evaluate its performance on medical image segmentation tasks from multiple perspectives, demonstrating superior accuracy and scalability compared to existing interactive segmentation models. To facilitate research on foundational models in medical computer vision, we release the IMed-361M and model at https://github.com/uni-medical/IMIS-Bench.
Efficient and generalizable nested Fourier-DeepONet for three-dimensional geological carbon sequestration
Sep 25, 2024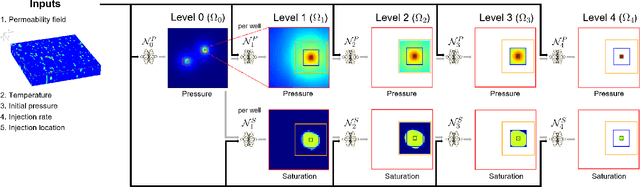
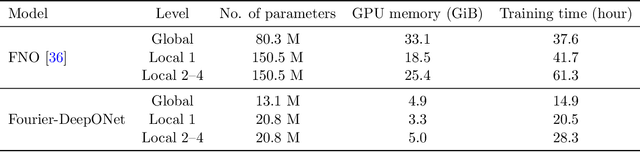
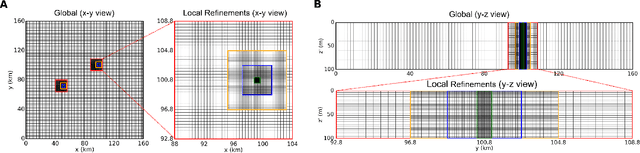
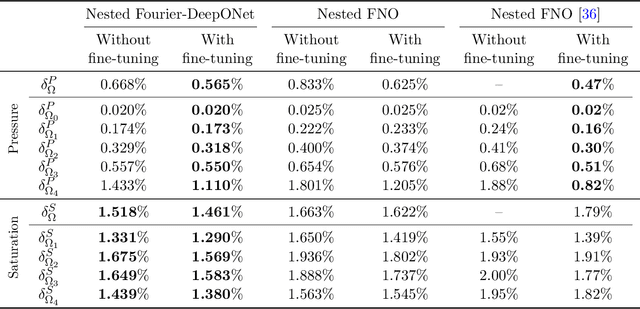
Abstract:Geological carbon sequestration (GCS) involves injecting CO$_2$ into subsurface geological formations for permanent storage. Numerical simulations could guide decisions in GCS projects by predicting CO$_2$ migration pathways and the pressure distribution in storage formation. However, these simulations are often computationally expensive due to highly coupled physics and large spatial-temporal simulation domains. Surrogate modeling with data-driven machine learning has become a promising alternative to accelerate physics-based simulations. Among these, the Fourier neural operator (FNO) has been applied to three-dimensional synthetic subsurface models. Here, to further improve performance, we have developed a nested Fourier-DeepONet by combining the expressiveness of the FNO with the modularity of a deep operator network (DeepONet). This new framework is twice as efficient as a nested FNO for training and has at least 80% lower GPU memory requirement due to its flexibility to treat temporal coordinates separately. These performance improvements are achieved without compromising prediction accuracy. In addition, the generalization and extrapolation ability of nested Fourier-DeepONet beyond the training range has been thoroughly evaluated. Nested Fourier-DeepONet outperformed the nested FNO for extrapolation in time with more than 50% reduced error. It also exhibited good extrapolation accuracy beyond the training range in terms of reservoir properties, number of wells, and injection rate.
Beyond First-Order: A Multi-Scale Approach to Finger Knuckle Print Biometrics
Jun 28, 2024Abstract:Recently, finger knuckle prints (FKPs) have gained attention due to their rich textural patterns, positioning them as a promising biometric for identity recognition. Prior FKP recognition methods predominantly leverage first-order feature descriptors, which capture intricate texture details but fail to account for structural information. Emerging research, however, indicates that second-order textures, which describe the curves and arcs of the textures, encompass this overlooked structural information. This paper introduces a novel FKP recognition approach, the Dual-Order Texture Competition Network (DOTCNet), designed to capture texture information in FKP images comprehensively. DOTCNet incorporates three dual-order texture competitive modules (DTCMs), each targeting textures at different scales. Each DTCM employs a learnable texture descriptor, specifically a learnable Gabor filter (LGF), to extract texture features. By leveraging LGFs, the network extracts first and second order textures to describe fine textures and structural features thoroughly. Furthermore, an attention mechanism enhances relevant features in the first-order features, thereby highlighting significant texture details. For second-order features, a competitive mechanism emphasizes structural information while reducing noise from higher-order features. Extensive experimental results reveal that DOTCNet significantly outperforms several standard algorithms on the publicly available PolyU-FKP dataset.
Cephalometric Landmark Detection across Ages with Prototypical Network
Jun 18, 2024



Abstract:Automated cephalometric landmark detection is crucial in real-world orthodontic diagnosis. Current studies mainly focus on only adult subjects, neglecting the clinically crucial scenario presented by adolescents whose landmarks often exhibit significantly different appearances compared to adults. Hence, an open question arises about how to develop a unified and effective detection algorithm across various age groups, including adolescents and adults. In this paper, we propose CeLDA, the first work for Cephalometric Landmark Detection across Ages. Our method leverages a prototypical network for landmark detection by comparing image features with landmark prototypes. To tackle the appearance discrepancy of landmarks between age groups, we design new strategies for CeLDA to improve prototype alignment and obtain a holistic estimation of landmark prototypes from a large set of training images. Moreover, a novel prototype relation mining paradigm is introduced to exploit the anatomical relations between the landmark prototypes. Extensive experiments validate the superiority of CeLDA in detecting cephalometric landmarks on both adult and adolescent subjects. To our knowledge, this is the first effort toward developing a unified solution and dataset for cephalometric landmark detection across age groups. Our code and dataset will be made public on https://github.com/ShanghaiTech-IMPACT/Cephalometric-Landmark-Detection-across-Ages-with-Prototypical-Network
Scale-aware competition network for palmprint recognition
Nov 21, 2023Abstract:Palmprint biometrics garner heightened attention in palm-scanning payment and social security due to their distinctive attributes. However, prevailing methodologies singularly prioritize texture orientation, neglecting the significant texture scale dimension. We design an innovative network for concurrently extracting intra-scale and inter-scale features to redress this limitation. This paper proposes a scale-aware competitive network (SAC-Net), which includes the Inner-Scale Competition Module (ISCM) and the Across-Scale Competition Module (ASCM) to capture texture characteristics related to orientation and scale. ISCM efficiently integrates learnable Gabor filters and a self-attention mechanism to extract rich orientation data and discern textures with long-range discriminative properties. Subsequently, ASCM leverages a competitive strategy across various scales to effectively encapsulate the competitive texture scale elements. By synergizing ISCM and ASCM, our method adeptly characterizes palmprint features. Rigorous experimentation across three benchmark datasets unequivocally demonstrates our proposed approach's exceptional recognition performance and resilience relative to state-of-the-art alternatives.
SA-Med2D-20M Dataset: Segment Anything in 2D Medical Imaging with 20 Million masks
Nov 20, 2023



Abstract:Segment Anything Model (SAM) has achieved impressive results for natural image segmentation with input prompts such as points and bounding boxes. Its success largely owes to massive labeled training data. However, directly applying SAM to medical image segmentation cannot perform well because SAM lacks medical knowledge -- it does not use medical images for training. To incorporate medical knowledge into SAM, we introduce SA-Med2D-20M, a large-scale segmentation dataset of 2D medical images built upon numerous public and private datasets. It consists of 4.6 million 2D medical images and 19.7 million corresponding masks, covering almost the whole body and showing significant diversity. This paper describes all the datasets collected in SA-Med2D-20M and details how to process these datasets. Furthermore, comprehensive statistics of SA-Med2D-20M are presented to facilitate the better use of our dataset, which can help the researchers build medical vision foundation models or apply their models to downstream medical applications. We hope that the large scale and diversity of SA-Med2D-20M can be leveraged to develop medical artificial intelligence for enhancing diagnosis, medical image analysis, knowledge sharing, and education. The data with the redistribution license is publicly available at https://github.com/OpenGVLab/SAM-Med2D.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge