Pradeep Singh
Benchmarking LLMs for Predictive Applications in the Intensive Care Units
Dec 23, 2025



Abstract:With the advent of LLMs, various tasks across the natural language processing domain have been transformed. However, their application in predictive tasks remains less researched. This study compares large language models, including GatorTron-Base (trained on clinical data), Llama 8B, and Mistral 7B, against models like BioBERT, DocBERT, BioClinicalBERT, Word2Vec, and Doc2Vec, setting benchmarks for predicting Shock in critically ill patients. Timely prediction of shock can enable early interventions, thus improving patient outcomes. Text data from 17,294 ICU stays of patients in the MIMIC III database were scored for length of stay > 24 hours and shock index (SI) > 0.7 to yield 355 and 87 patients with normal and abnormal SI-index, respectively. Both focal and cross-entropy losses were used during finetuning to address class imbalances. Our findings indicate that while GatorTron Base achieved the highest weighted recall of 80.5%, the overall performance metrics were comparable between SLMs and LLMs. This suggests that LLMs are not inherently superior to SLMs in predicting future clinical events despite their strong performance on text-based tasks. To achieve meaningful clinical outcomes, future efforts in training LLMs should prioritize developing models capable of predicting clinical trajectories rather than focusing on simpler tasks such as named entity recognition or phenotyping.
The Universe Learning Itself: On the Evolution of Dynamics from the Big Bang to Machine Intelligence
Dec 21, 2025Abstract:We develop a unified, dynamical-systems narrative of the universe that traces a continuous chain of structure formation from the Big Bang to contemporary human societies and their artificial learning systems. Rather than treating cosmology, astrophysics, geophysics, biology, cognition, and machine intelligence as disjoint domains, we view each as successive regimes of dynamics on ever-richer state spaces, stitched together by phase transitions, symmetry-breaking events, and emergent attractors. Starting from inflationary field dynamics and the growth of primordial perturbations, we describe how gravitational instability sculpts the cosmic web, how dissipative collapse in baryonic matter yields stars and planets, and how planetary-scale geochemical cycles define long-lived nonequilibrium attractors. Within these attractors, we frame the origin of life as the emergence of self-maintaining reaction networks, evolutionary biology as flow on high-dimensional genotype-phenotype-environment manifolds, and brains as adaptive dynamical systems operating near critical surfaces. Human culture and technology-including modern machine learning and artificial intelligence-are then interpreted as symbolic and institutional dynamics that implement and refine engineered learning flows which recursively reshape their own phase space. Throughout, we emphasize recurring mathematical motifs-instability, bifurcation, multiscale coupling, and constrained flows on measure-zero subsets of the accessible state space. Our aim is not to present any new cosmological or biological model, but a cross-scale, theoretical perspective: a way of reading the universe's history as the evolution of dynamics itself, culminating (so far) in biological and artificial systems capable of modeling, predicting, and deliberately perturbing their own future trajectories.
Echo State Networks as State-Space Models: A Systems Perspective
Sep 04, 2025Abstract:Echo State Networks (ESNs) are typically presented as efficient, readout-trained recurrent models, yet their dynamics and design are often guided by heuristics rather than first principles. We recast ESNs explicitly as state-space models (SSMs), providing a unified systems-theoretic account that links reservoir computing with classical identification and modern kernelized SSMs. First, we show that the echo-state property is an instance of input-to-state stability for a contractive nonlinear SSM and derive verifiable conditions in terms of leak, spectral scaling, and activation Lipschitz constants. Second, we develop two complementary mappings: (i) small-signal linearizations that yield locally valid LTI SSMs with interpretable poles and memory horizons; and (ii) lifted/Koopman random-feature expansions that render the ESN a linear SSM in an augmented state, enabling transfer-function and convolutional-kernel analyses. This perspective yields frequency-domain characterizations of memory spectra and clarifies when ESNs emulate structured SSM kernels. Third, we cast teacher forcing as state estimation and propose Kalman/EKF-assisted readout learning, together with EM for hyperparameters (leak, spectral radius, process/measurement noise) and a hybrid subspace procedure for spectral shaping under contraction constraints.
Frozen in Time: Parameter-Efficient Time Series Transformers via Reservoir-Induced Feature Expansion and Fixed Random Dynamics
Aug 25, 2025Abstract:Transformers are the de-facto choice for sequence modelling, yet their quadratic self-attention and weak temporal bias can make long-range forecasting both expensive and brittle. We introduce FreezeTST, a lightweight hybrid that interleaves frozen random-feature (reservoir) blocks with standard trainable Transformer layers. The frozen blocks endow the network with rich nonlinear memory at no optimisation cost; the trainable layers learn to query this memory through self-attention. The design cuts trainable parameters and also lowers wall-clock training time, while leaving inference complexity unchanged. On seven standard long-term forecasting benchmarks, FreezeTST consistently matches or surpasses specialised variants such as Informer, Autoformer, and PatchTST; with substantially lower compute. Our results show that embedding reservoir principles within Transformers offers a simple, principled route to efficient long-term time-series prediction.
HypER: Hyperbolic Echo State Networks for Capturing Stretch-and-Fold Dynamics in Chaotic Flows
Aug 25, 2025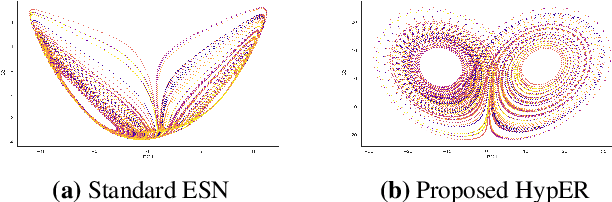
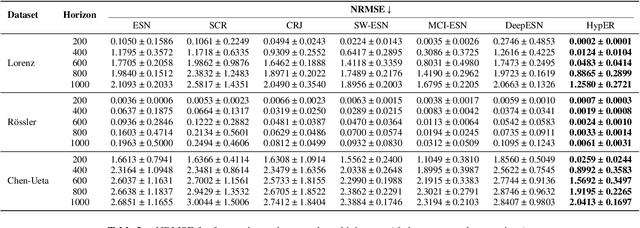
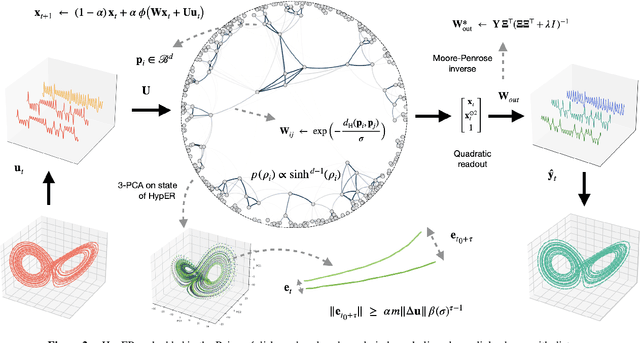
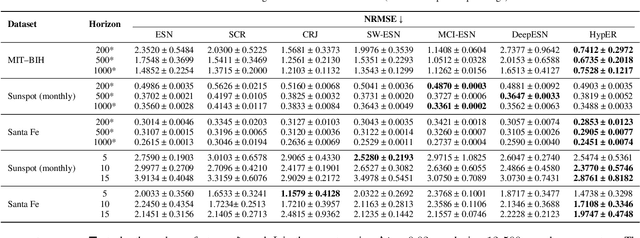
Abstract:Forecasting chaotic dynamics beyond a few Lyapunov times is difficult because infinitesimal errors grow exponentially. Existing Echo State Networks (ESNs) mitigate this growth but employ reservoirs whose Euclidean geometry is mismatched to the stretch-and-fold structure of chaos. We introduce the Hyperbolic Embedding Reservoir (HypER), an ESN whose neurons are sampled in the Poincare ball and whose connections decay exponentially with hyperbolic distance. This negative-curvature construction embeds an exponential metric directly into the latent space, aligning the reservoir's local expansion-contraction spectrum with the system's Lyapunov directions while preserving standard ESN features such as sparsity, leaky integration, and spectral-radius control. Training is limited to a Tikhonov-regularized readout. On the chaotic Lorenz-63 and Roessler systems, and the hyperchaotic Chen-Ueta attractor, HypER consistently lengthens the mean valid-prediction horizon beyond Euclidean and graph-structured ESN baselines, with statistically significant gains confirmed over 30 independent runs; parallel results on real-world benchmarks, including heart-rate variability from the Santa Fe and MIT-BIH datasets and international sunspot numbers, corroborate its advantage. We further establish a lower bound on the rate of state divergence for HypER, mirroring Lyapunov growth.
Dynamics and Computational Principles of Echo State Networks: A Mathematical Perspective
Apr 16, 2025Abstract:Reservoir computing (RC) represents a class of state-space models (SSMs) characterized by a fixed state transition mechanism (the reservoir) and a flexible readout layer that maps from the state space. It is a paradigm of computational dynamical systems that harnesses the transient dynamics of high-dimensional state spaces for efficient processing of temporal data. Rooted in concepts from recurrent neural networks, RC achieves exceptional computational power by decoupling the training of the dynamic reservoir from the linear readout layer, thereby circumventing the complexities of gradient-based optimization. This work presents a systematic exploration of RC, addressing its foundational properties such as the echo state property, fading memory, and reservoir capacity through the lens of dynamical systems theory. We formalize the interplay between input signals and reservoir states, demonstrating the conditions under which reservoirs exhibit stability and expressive power. Further, we delve into the computational trade-offs and robustness characteristics of RC architectures, extending the discussion to their applications in signal processing, time-series prediction, and control systems. The analysis is complemented by theoretical insights into optimization, training methodologies, and scalability, highlighting open challenges and potential directions for advancing the theoretical underpinnings of RC.
Training-free Quantum-Inspired Image Edge Extraction Method
Jan 31, 2025Abstract:Edge detection is a cornerstone of image processing, yet existing methods often face critical limitations. Traditional deep learning edge detection methods require extensive training datasets and fine-tuning, while classical techniques often fail in complex or noisy scenarios, limiting their real-world applicability. To address these limitations, we propose a training-free, quantum-inspired edge detection model. Our approach integrates classical Sobel edge detection, the Schr\"odinger wave equation refinement, and a hybrid framework combining Canny and Laplacian operators. By eliminating the need for training, the model is lightweight and adaptable to diverse applications. The Schr\"odinger wave equation refines gradient-based edge maps through iterative diffusion, significantly enhancing edge precision. The hybrid framework further strengthens the model by synergistically combining local and global features, ensuring robustness even under challenging conditions. Extensive evaluations on datasets like BIPED, Multicue, and NYUD demonstrate superior performance of the proposed model, achieving state-of-the-art metrics, including ODS, OIS, AP, and F-measure. Noise robustness experiments highlight its reliability, showcasing its practicality for real-world scenarios. Due to its versatile and adaptable nature, our model is well-suited for applications such as medical imaging, autonomous systems, and environmental monitoring, setting a new benchmark for edge detection.
Facial Surgery Preview Based on the Orthognathic Treatment Prediction
Dec 15, 2024



Abstract:Orthognathic surgery consultation is essential to help patients understand the changes to their facial appearance after surgery. However, current visualization methods are often inefficient and inaccurate due to limited pre- and post-treatment data and the complexity of the treatment. To overcome these challenges, this study aims to develop a fully automated pipeline that generates accurate and efficient 3D previews of postsurgical facial appearances for patients with orthognathic treatment without requiring additional medical images. The study introduces novel aesthetic losses, such as mouth-convexity and asymmetry losses, to improve the accuracy of facial surgery prediction. Additionally, it proposes a specialized parametric model for 3D reconstruction of the patient, medical-related losses to guide latent code prediction network optimization, and a data augmentation scheme to address insufficient data. The study additionally employs FLAME, a parametric model, to enhance the quality of facial appearance previews by extracting facial latent codes and establishing dense correspondences between pre- and post-surgery geometries. Quantitative comparisons showed the algorithm's effectiveness, and qualitative results highlighted accurate facial contour and detail predictions. A user study confirmed that doctors and the public could not distinguish between machine learning predictions and actual postoperative results. This study aims to offer a practical, effective solution for orthognathic surgery consultations, benefiting doctors and patients.
DeepMediX: A Deep Learning-Driven Resource-Efficient Medical Diagnosis Across the Spectrum
Jul 01, 2023



Abstract:In the rapidly evolving landscape of medical imaging diagnostics, achieving high accuracy while preserving computational efficiency remains a formidable challenge. This work presents \texttt{DeepMediX}, a groundbreaking, resource-efficient model that significantly addresses this challenge. Built on top of the MobileNetV2 architecture, DeepMediX excels in classifying brain MRI scans and skin cancer images, with superior performance demonstrated on both binary and multiclass skin cancer datasets. It provides a solution to labor-intensive manual processes, the need for large datasets, and complexities related to image properties. DeepMediX's design also includes the concept of Federated Learning, enabling a collaborative learning approach without compromising data privacy. This approach allows diverse healthcare institutions to benefit from shared learning experiences without the necessity of direct data access, enhancing the model's predictive power while preserving the privacy and integrity of sensitive patient data. Its low computational footprint makes DeepMediX suitable for deployment on handheld devices, offering potential for real-time diagnostic support. Through rigorous testing on standard datasets, including the ISIC2018 for dermatological research, DeepMediX demonstrates exceptional diagnostic capabilities, matching the performance of existing models on almost all tasks and even outperforming them in some cases. The findings of this study underline significant implications for the development and deployment of AI-based tools in medical imaging and their integration into point-of-care settings. The source code and models generated would be released at https://github.com/kishorebabun/DeepMediX.
See Through the Fog: Curriculum Learning with Progressive Occlusion in Medical Imaging
Jun 30, 2023Abstract:In recent years, deep learning models have revolutionized medical image interpretation, offering substantial improvements in diagnostic accuracy. However, these models often struggle with challenging images where critical features are partially or fully occluded, which is a common scenario in clinical practice. In this paper, we propose a novel curriculum learning-based approach to train deep learning models to handle occluded medical images effectively. Our method progressively introduces occlusion, starting from clear, unobstructed images and gradually moving to images with increasing occlusion levels. This ordered learning process, akin to human learning, allows the model to first grasp simple, discernable patterns and subsequently build upon this knowledge to understand more complicated, occluded scenarios. Furthermore, we present three novel occlusion synthesis methods, namely Wasserstein Curriculum Learning (WCL), Information Adaptive Learning (IAL), and Geodesic Curriculum Learning (GCL). Our extensive experiments on diverse medical image datasets demonstrate substantial improvements in model robustness and diagnostic accuracy over conventional training methodologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge