Kishore Babu Nampalle
DeepMediX: A Deep Learning-Driven Resource-Efficient Medical Diagnosis Across the Spectrum
Jul 01, 2023



Abstract:In the rapidly evolving landscape of medical imaging diagnostics, achieving high accuracy while preserving computational efficiency remains a formidable challenge. This work presents \texttt{DeepMediX}, a groundbreaking, resource-efficient model that significantly addresses this challenge. Built on top of the MobileNetV2 architecture, DeepMediX excels in classifying brain MRI scans and skin cancer images, with superior performance demonstrated on both binary and multiclass skin cancer datasets. It provides a solution to labor-intensive manual processes, the need for large datasets, and complexities related to image properties. DeepMediX's design also includes the concept of Federated Learning, enabling a collaborative learning approach without compromising data privacy. This approach allows diverse healthcare institutions to benefit from shared learning experiences without the necessity of direct data access, enhancing the model's predictive power while preserving the privacy and integrity of sensitive patient data. Its low computational footprint makes DeepMediX suitable for deployment on handheld devices, offering potential for real-time diagnostic support. Through rigorous testing on standard datasets, including the ISIC2018 for dermatological research, DeepMediX demonstrates exceptional diagnostic capabilities, matching the performance of existing models on almost all tasks and even outperforming them in some cases. The findings of this study underline significant implications for the development and deployment of AI-based tools in medical imaging and their integration into point-of-care settings. The source code and models generated would be released at https://github.com/kishorebabun/DeepMediX.
See Through the Fog: Curriculum Learning with Progressive Occlusion in Medical Imaging
Jun 30, 2023Abstract:In recent years, deep learning models have revolutionized medical image interpretation, offering substantial improvements in diagnostic accuracy. However, these models often struggle with challenging images where critical features are partially or fully occluded, which is a common scenario in clinical practice. In this paper, we propose a novel curriculum learning-based approach to train deep learning models to handle occluded medical images effectively. Our method progressively introduces occlusion, starting from clear, unobstructed images and gradually moving to images with increasing occlusion levels. This ordered learning process, akin to human learning, allows the model to first grasp simple, discernable patterns and subsequently build upon this knowledge to understand more complicated, occluded scenarios. Furthermore, we present three novel occlusion synthesis methods, namely Wasserstein Curriculum Learning (WCL), Information Adaptive Learning (IAL), and Geodesic Curriculum Learning (GCL). Our extensive experiments on diverse medical image datasets demonstrate substantial improvements in model robustness and diagnostic accuracy over conventional training methodologies.
Vision Through the Veil: Differential Privacy in Federated Learning for Medical Image Classification
Jun 30, 2023Abstract:The proliferation of deep learning applications in healthcare calls for data aggregation across various institutions, a practice often associated with significant privacy concerns. This concern intensifies in medical image analysis, where privacy-preserving mechanisms are paramount due to the data being sensitive in nature. Federated learning, which enables cooperative model training without direct data exchange, presents a promising solution. Nevertheless, the inherent vulnerabilities of federated learning necessitate further privacy safeguards. This study addresses this need by integrating differential privacy, a leading privacy-preserving technique, into a federated learning framework for medical image classification. We introduce a novel differentially private federated learning model and meticulously examine its impacts on privacy preservation and model performance. Our research confirms the existence of a trade-off between model accuracy and privacy settings. However, we demonstrate that strategic calibration of the privacy budget in differential privacy can uphold robust image classification performance while providing substantial privacy protection.
Transcending Grids: Point Clouds and Surface Representations Powering Neurological Processing
May 17, 2023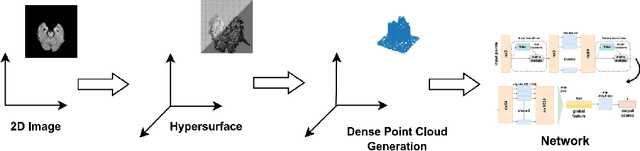
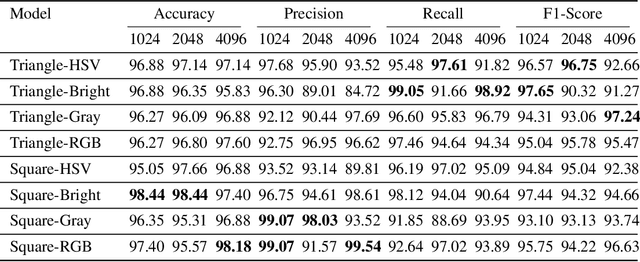
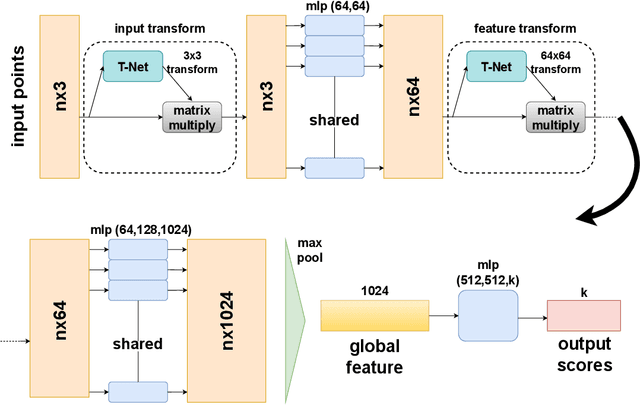
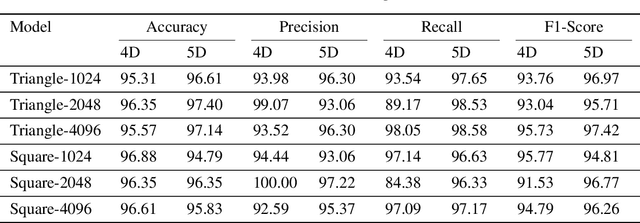
Abstract:In healthcare, accurately classifying medical images is vital, but conventional methods often hinge on medical data with a consistent grid structure, which may restrict their overall performance. Recent medical research has been focused on tweaking the architectures to attain better performance without giving due consideration to the representation of data. In this paper, we present a novel approach for transforming grid based data into its higher dimensional representations, leveraging unstructured point cloud data structures. We first generate a sparse point cloud from an image by integrating pixel color information as spatial coordinates. Next, we construct a hypersurface composed of points based on the image dimensions, with each smooth section within this hypersurface symbolizing a specific pixel location. Polygonal face construction is achieved using an adjacency tensor. Finally, a dense point cloud is generated by densely sampling the constructed hypersurface, with a focus on regions of higher detail. The effectiveness of our approach is demonstrated on a publicly accessible brain tumor dataset, achieving significant improvements over existing classification techniques. This methodology allows the extraction of intricate details from the original image, opening up new possibilities for advanced image analysis and processing tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge