Linrui Dai
CT Synthesis with Conditional Diffusion Models for Abdominal Lymph Node Segmentation
Mar 26, 2024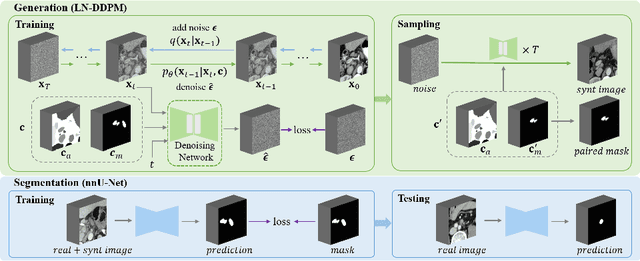
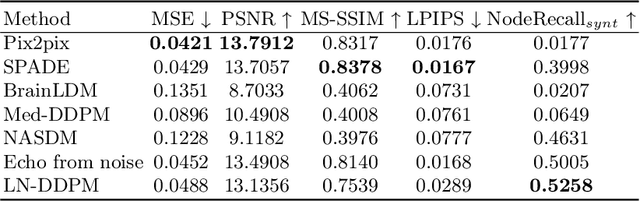
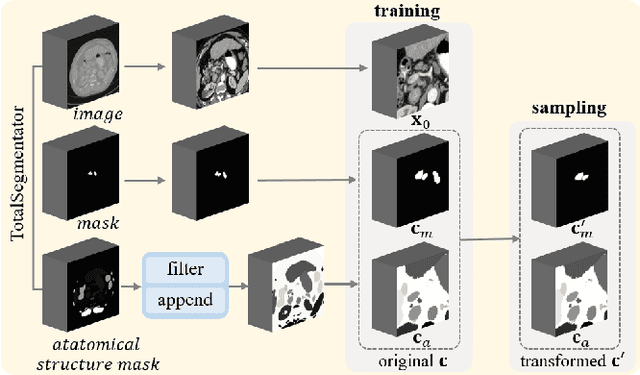
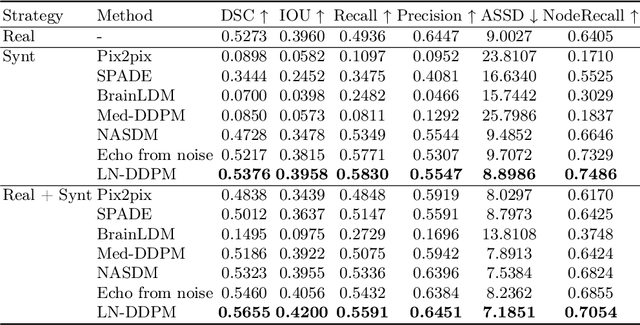
Abstract:Despite the significant success achieved by deep learning methods in medical image segmentation, researchers still struggle in the computer-aided diagnosis of abdominal lymph nodes due to the complex abdominal environment, small and indistinguishable lesions, and limited annotated data. To address these problems, we present a pipeline that integrates the conditional diffusion model for lymph node generation and the nnU-Net model for lymph node segmentation to improve the segmentation performance of abdominal lymph nodes through synthesizing a diversity of realistic abdominal lymph node data. We propose LN-DDPM, a conditional denoising diffusion probabilistic model (DDPM) for lymph node (LN) generation. LN-DDPM utilizes lymph node masks and anatomical structure masks as model conditions. These conditions work in two conditioning mechanisms: global structure conditioning and local detail conditioning, to distinguish between lymph nodes and their surroundings and better capture lymph node characteristics. The obtained paired abdominal lymph node images and masks are used for the downstream segmentation task. Experimental results on the abdominal lymph node datasets demonstrate that LN-DDPM outperforms other generative methods in the abdominal lymph node image synthesis and better assists the downstream abdominal lymph node segmentation task.
GuideGen: A Text-guided Framework for Joint CT Volume and Anatomical structure Generation
Mar 12, 2024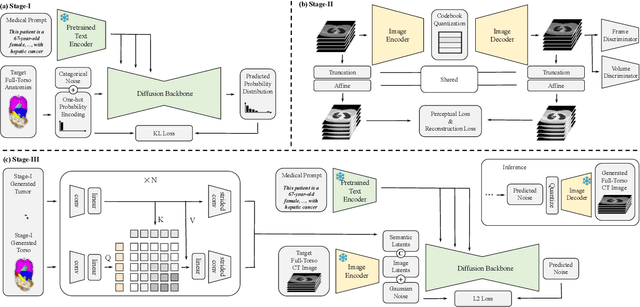
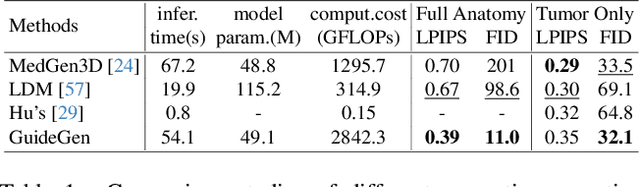
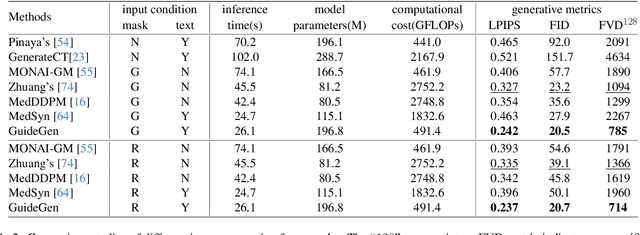
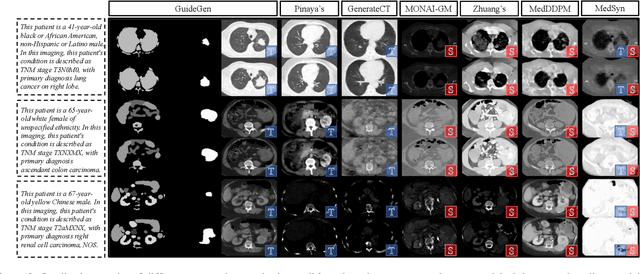
Abstract:The annotation burden and extensive labor for gathering a large medical dataset with images and corresponding labels are rarely cost-effective and highly intimidating. This results in a lack of abundant training data that undermines downstream tasks and partially contributes to the challenge image analysis faces in the medical field. As a workaround, given the recent success of generative neural models, it is now possible to synthesize image datasets at a high fidelity guided by external constraints. This paper explores this possibility and presents \textbf{GuideGen}: a pipeline that jointly generates CT images and tissue masks for abdominal organs and colorectal cancer conditioned on a text prompt. Firstly, we introduce Volumetric Mask Sampler to fit the discrete distribution of mask labels and generate low-resolution 3D tissue masks. Secondly, our Conditional Image Generator autoregressively generates CT slices conditioned on a corresponding mask slice to incorporate both style information and anatomical guidance. This pipeline guarantees high fidelity and variability as well as exact alignment between generated CT volumes and tissue masks. Both qualitative and quantitative experiments on 3D abdominal CTs demonstrate a high performance of our proposed pipeline, thereby proving our method can serve as a dataset generator and provide potential benefits to downstream tasks. It is hoped that our work will offer a promising solution on the multimodality generation of CT and its anatomical mask. Our source code is publicly available at https://github.com/OvO1111/JointImageGeneration.
Efficient Subclass Segmentation in Medical Images
Jul 01, 2023



Abstract:As research interests in medical image analysis become increasingly fine-grained, the cost for extensive annotation also rises. One feasible way to reduce the cost is to annotate with coarse-grained superclass labels while using limited fine-grained annotations as a complement. In this way, fine-grained data learning is assisted by ample coarse annotations. Recent studies in classification tasks have adopted this method to achieve satisfactory results. However, there is a lack of research on efficient learning of fine-grained subclasses in semantic segmentation tasks. In this paper, we propose a novel approach that leverages the hierarchical structure of categories to design network architecture. Meanwhile, a task-driven data generation method is presented to make it easier for the network to recognize different subclass categories. Specifically, we introduce a Prior Concatenation module that enhances confidence in subclass segmentation by concatenating predicted logits from the superclass classifier, a Separate Normalization module that stretches the intra-class distance within the same superclass to facilitate subclass segmentation, and a HierarchicalMix model that generates high-quality pseudo labels for unlabeled samples by fusing only similar superclass regions from labeled and unlabeled images. Our experiments on the BraTS2021 and ACDC datasets demonstrate that our approach achieves comparable accuracy to a model trained with full subclass annotations, with limited subclass annotations and sufficient superclass annotations. Our approach offers a promising solution for efficient fine-grained subclass segmentation in medical images. Our code is publicly available here.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge