Louise Piecuch
MD
Implicit Shape-Prior for Few-Shot Assisted 3D Segmentation
Sep 10, 2025Abstract:The objective of this paper is to significantly reduce the manual workload required from medical professionals in complex 3D segmentation tasks that cannot be yet fully automated. For instance, in radiotherapy planning, organs at risk must be accurately identified in computed tomography (CT) or magnetic resonance imaging (MRI) scans to ensure they are spared from harmful radiation. Similarly, diagnosing age-related degenerative diseases such as sarcopenia, which involve progressive muscle volume loss and strength, is commonly based on muscular mass measurements often obtained from manual segmentation of medical volumes. To alleviate the manual-segmentation burden, this paper introduces an implicit shape prior to segment volumes from sparse slice manual annotations generalized to the multi-organ case, along with a simple framework for automatically selecting the most informative slices to guide and minimize the next interactions. The experimental validation shows the method's effectiveness on two medical use cases: assisted segmentation in the context of at risks organs for brain cancer patients, and acceleration of the creation of a new database with unseen muscle shapes for patients with sarcopenia.
Unsupervised Anomaly Detection on Implicit Shape representations for Sarcopenia Detection
Feb 13, 2025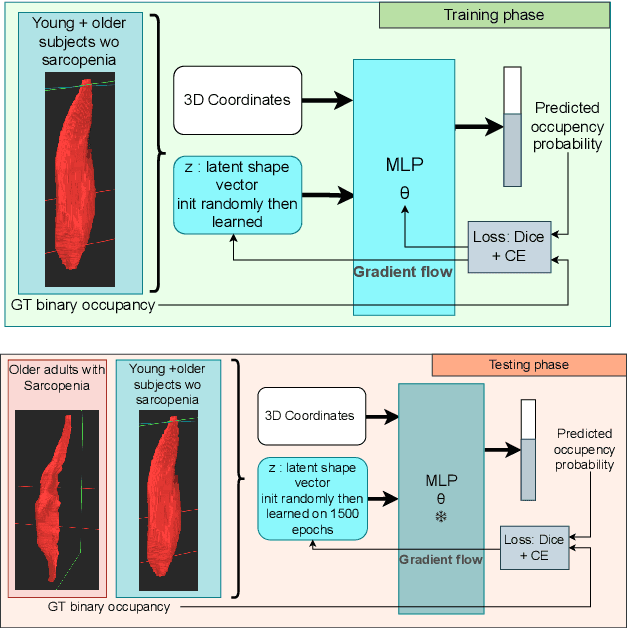
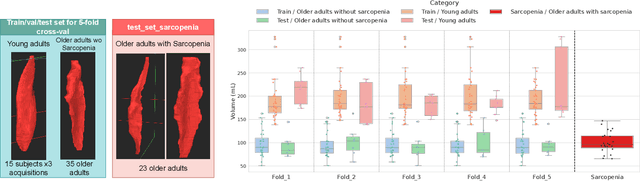
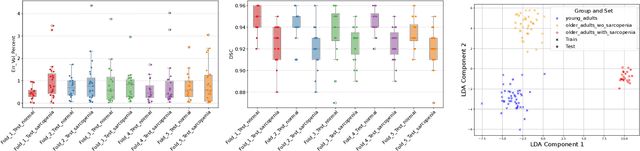
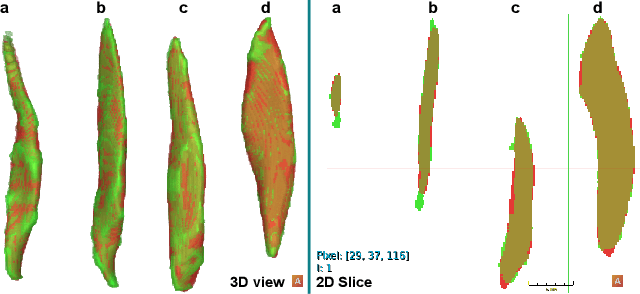
Abstract:Sarcopenia is an age-related progressive loss of muscle mass and strength that significantly impacts daily life. A commonly studied criterion for characterizing the muscle mass has been the combination of 3D imaging and manual segmentations. In this paper, we instead study the muscles' shape. We rely on an implicit neural representation (INR) to model normal muscle shapes. We then introduce an unsupervised anomaly detection method to identify sarcopenic muscles based on the reconstruction error of the implicit model. Relying on a conditional INR with an auto-decoding strategy, we also learn a latent representation of the muscles that clearly separates normal from abnormal muscles in an unsupervised fashion. Experimental results on a dataset of 103 segmented volumes indicate that our double anomaly detection strategy effectively discriminates sarcopenic and non-sarcopenic muscles.
Muscle volume quantification: guiding transformers with anatomical priors
Oct 31, 2023Abstract:Muscle volume is a useful quantitative biomarker in sports, but also for the follow-up of degenerative musculo-skelletal diseases. In addition to volume, other shape biomarkers can be extracted by segmenting the muscles of interest from medical images. Manual segmentation is still today the gold standard for such measurements despite being very time-consuming. We propose a method for automatic segmentation of 18 muscles of the lower limb on 3D Magnetic Resonance Images to assist such morphometric analysis. By their nature, the tissue of different muscles is undistinguishable when observed in MR Images. Thus, muscle segmentation algorithms cannot rely on appearance but only on contour cues. However, such contours are hard to detect and their thickness varies across subjects. To cope with the above challenges, we propose a segmentation approach based on a hybrid architecture, combining convolutional and visual transformer blocks. We investigate for the first time the behaviour of such hybrid architectures in the context of muscle segmentation for shape analysis. Considering the consistent anatomical muscle configuration, we rely on transformer blocks to capture the longrange relations between the muscles. To further exploit the anatomical priors, a second contribution of this work consists in adding a regularisation loss based on an adjacency matrix of plausible muscle neighbourhoods estimated from the training data. Our experimental results on a unique database of elite athletes show it is possible to train complex hybrid models from a relatively small database of large volumes, while the anatomical prior regularisation favours better predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge