Gabriel Bernardino
High-dimensional multimodal uncertainty estimation by manifold alignment:Application to 3D right ventricular strain computations
Jan 21, 2025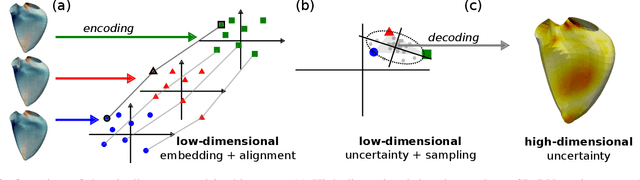
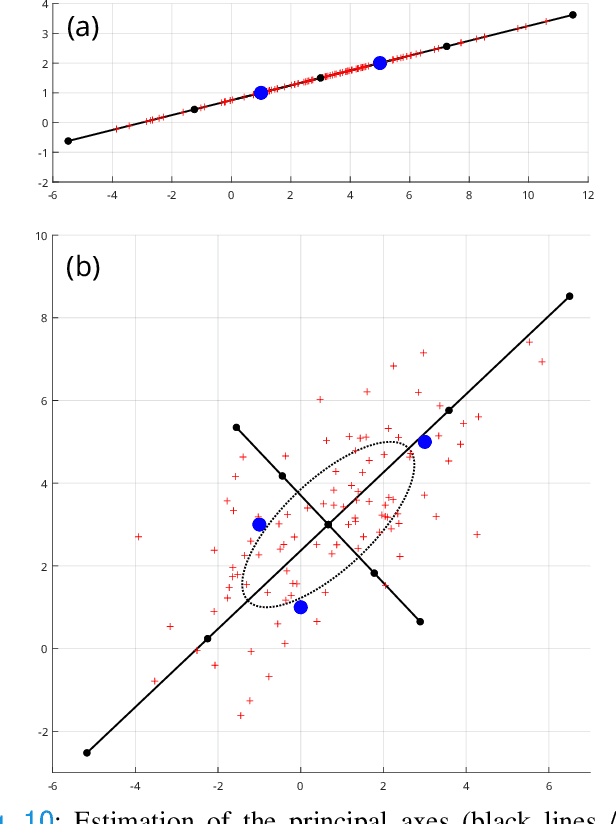
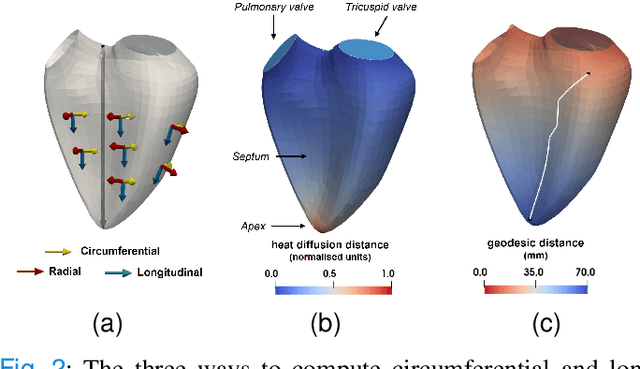
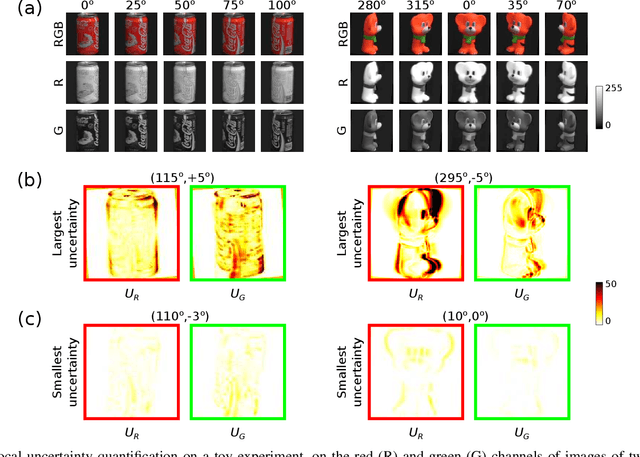
Abstract:Confidence in the results is a key ingredient to improve the adoption of machine learning methods by clinicians. Uncertainties on the results have been considered in the literature, but mostly those originating from the learning and processing methods. Uncertainty on the data is hardly challenged, as a single sample is often considered representative enough of each subject included in the analysis. In this paper, we propose a representation learning strategy to estimate local uncertainties on a physiological descriptor (here, myocardial deformation) previously obtained from medical images by different definitions or computations. We first use manifold alignment to match the latent representations associated to different high-dimensional input descriptors. Then, we formulate plausible distributions of latent uncertainties, and finally exploit them to reconstruct uncertainties on the input high-dimensional descriptors. We demonstrate its relevance for the quantification of myocardial deformation (strain) from 3D echocardiographic image sequences of the right ventricle, for which a lack of consensus exists in its definition and which directional component to use. We used a database of 100 control subjects with right ventricle overload, for which different types of strain are available at each point of the right ventricle endocardial surface mesh. Our approach quantifies local uncertainties on myocardial deformation from different descriptors defining this physiological concept. Such uncertainties cannot be directly estimated by local statistics on such descriptors, potentially of heterogeneous types. Beyond this controlled illustrative application, our methodology has the potential to be generalized to many other population analyses considering heterogeneous high-dimensional descriptors.
Handling confounding variables in statistical shape analysis -- application to cardiac remodelling
Jul 28, 2020
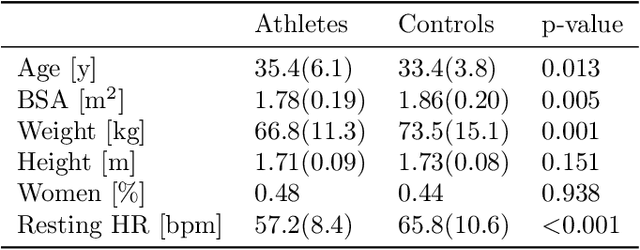
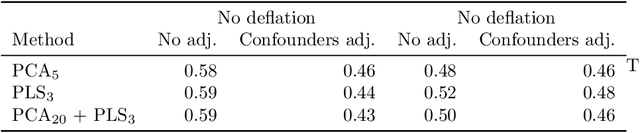
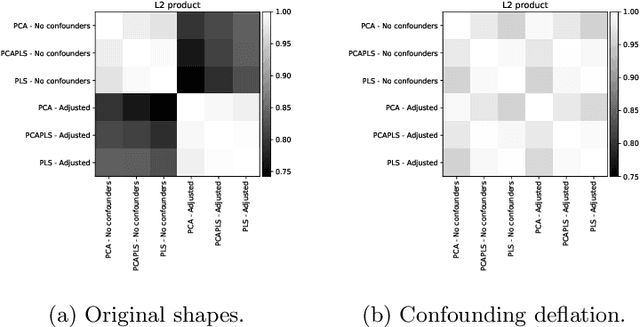
Abstract:Statistical shape analysis is a powerful tool to assess organ morphologies and find shape changes associated to a particular disease. However, imbalance in confounding factors, such as demographics might invalidate the analysis if not taken into consideration. Despite the methodological advances in the field, providing new methods that are able to capture complex and regional shape differences, the relationship between non-imaging information and shape variability has been overlooked. We present a linear statistical shape analysis framework that finds shape differences unassociated to a controlled set of confounding variables. It includes two confounding correction methods: confounding deflation and adjustment. We applied our framework to a cardiac magnetic resonance imaging dataset, consisting of the cardiac ventricles of 89 triathletes and 77 controls, to identify cardiac remodelling due to the practice of endurance exercise. To test robustness to confounders, subsets of this dataset were generated by randomly removing controls with low body mass index, thus introducing imbalance. The analysis of the whole dataset indicates an increase of ventricular volumes and myocardial mass in athletes, which is consistent with the clinical literature. However, when confounders are not taken into consideration no increase of myocardial mass is found. Using the downsampled datasets, we find that confounder adjustment methods are needed to find the real remodelling patterns in imbalanced datasets.
Volumetric parcellation of the right ventricle for regional geometric and functional assessment
Mar 18, 2020



Abstract:In clinical practice, assessment of right ventricle (RV) is primarily done through its global volume, given it is a standardised measurement, and has a good reproducibility in 3D modalities such as MRI and 3D echocardiography. However, many illness produce regionalchanges and therefore a local analysis could provide a better tool for understanding and diagnosis of illnesses. Current regional clinical measurements are 2D linear dimensions, and suffer of low reproducibility due to the difficulty to identify landmarks in the RV, specially in echocardiographic images due to its noise and artefacts. We proposed an automatic method for parcellating the RV cavity and compute regional volumes and ejection fractions in three regions: apex, inlet and outflow. We tested the reproducibility in 3D echocardiographic images. We also present a synthetic mesh-deformation method to generate a groundtruth dataset for validating the ability of the method to capture different types of remodelling. Results showed an acceptable intra-observer reproduciblity (<10%) but a higher inter-observer(>10%). The synthetic dataset allowed to identify that the method was capable of assessing global dilatations, and local dilatations in the circumferential direction but not longitudinal elongations
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge