Maxime Di Folco
Measuring and Aligning Abstraction in Vision-Language Models with Medical Taxonomies
Jan 21, 2026Abstract:Vision-Language Models show strong zero-shot performance for chest X-ray classification, but standard flat metrics fail to distinguish between clinically minor and severe errors. This work investigates how to quantify and mitigate abstraction errors by leveraging medical taxonomies. We benchmark several state-of-the-art VLMs using hierarchical metrics and introduce Catastrophic Abstraction Errors to capture cross-branch mistakes. Our results reveal substantial misalignment of VLMs with clinical taxonomies despite high flat performance. To address this, we propose risk-constrained thresholding and taxonomy-aware fine-tuning with radial embeddings, which reduce severe abstraction errors to below 2 per cent while maintaining competitive performance. These findings highlight the importance of hierarchical evaluation and representation-level alignment for safer and more clinically meaningful deployment of VLMs.
No Data? No Problem: Robust Vision-Tabular Learning with Missing Values
Dec 22, 2025



Abstract:Large-scale medical biobanks provide imaging data complemented by extensive tabular information, such as demographics or clinical measurements. However, this abundance of tabular attributes does not reflect real-world datasets, where only a subset of attributes may be available. This discrepancy calls for methods that can leverage all the tabular data during training while remaining robust to missing values at inference. To address this challenge, we propose RoVTL (Robust Vision-Tabular Learning), a framework designed to handle any level of tabular data availability, from 0% to 100%. RoVTL comprises two key stages: contrastive pretraining, where we introduce tabular attribute missingness as data augmentation to promote robustness, and downstream task tuning using a gated cross-attention module for multimodal fusion. During fine-tuning, we employ a novel Tabular More vs. Fewer loss that ranks performance based on the amount of available tabular data. Combined with disentangled gradient learning, this enables consistent performance across all tabular data completeness scenarios. We evaluate RoVTL on cardiac MRI scans from the UK Biobank, demonstrating superior robustness to missing tabular data compared to prior methods. Furthermore, RoVTL successfully generalizes to an external cardiac MRI dataset for multimodal disease classification, and extends to the natural images domain, achieving robust performance on a car advertisements dataset. The code is available at https://github.com/marteczkah/RoVTL.
Covariance Descriptors Meet General Vision Encoders: Riemannian Deep Learning for Medical Image Classification
Nov 06, 2025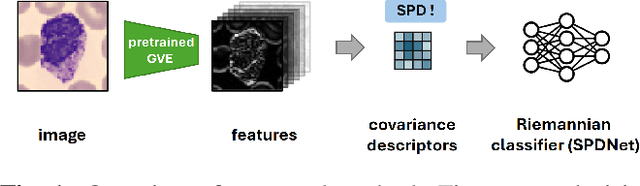
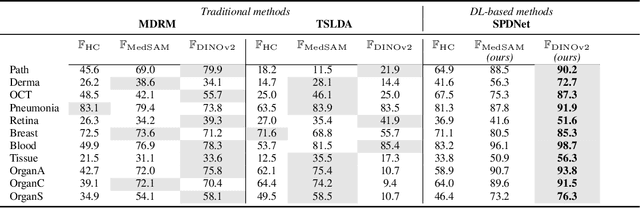
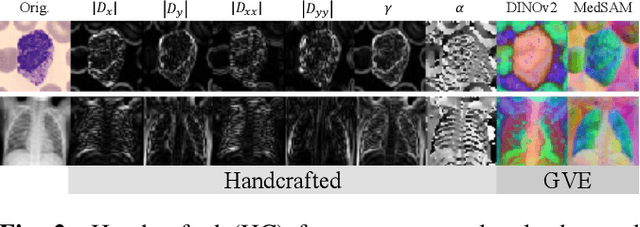
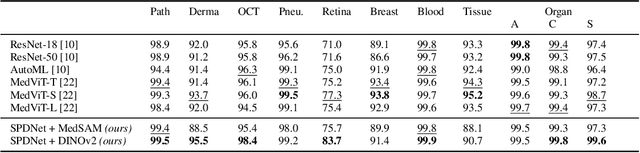
Abstract:Covariance descriptors capture second-order statistics of image features. They have shown strong performance in general computer vision tasks, but remain underexplored in medical imaging. We investigate their effectiveness for both conventional and learning-based medical image classification, with a particular focus on SPDNet, a classification network specifically designed for symmetric positive definite (SPD) matrices. We propose constructing covariance descriptors from features extracted by pre-trained general vision encoders (GVEs) and comparing them with handcrafted descriptors. Two GVEs - DINOv2 and MedSAM - are evaluated across eleven binary and multi-class datasets from the MedMNSIT benchmark. Our results show that covariance descriptors derived from GVE features consistently outperform those derived from handcrafted features. Moreover, SPDNet yields superior performance to state-of-the-art methods when combined with DINOv2 features. Our findings highlight the potential of combining covariance descriptors with powerful pretrained vision encoders for medical image analysis.
TGV: Tabular Data-Guided Learning of Visual Cardiac Representations
Mar 19, 2025Abstract:Contrastive learning methods in computer vision typically rely on different views of the same image to form pairs. However, in medical imaging, we often seek to compare entire patients with different phenotypes rather than just multiple augmentations of one scan. We propose harnessing clinically relevant tabular data to identify distinct patient phenotypes and form more meaningful pairs in a contrastive learning framework. Our method uses tabular attributes to guide the training of visual representations, without requiring a joint embedding space. We demonstrate its strength using short-axis cardiac MR images and clinical attributes from the UK Biobank, where tabular data helps to more effectively distinguish between patient subgroups. Evaluation on downstream tasks, including fine-tuning and zero-shot prediction of cardiovascular artery diseases and cardiac phenotypes, shows that incorporating tabular data yields stronger visual representations than conventional methods that rely solely on image augmentations or combined image-tabular embeddings. Furthermore, we demonstrate that image encoders trained with tabular guidance are capable of embedding demographic information in their representations, allowing them to use insights from tabular data for unimodal predictions, making them well-suited to real-world medical settings where extensive clinical annotations may not be routinely available at inference time. The code will be available on GitHub.
Semantic Alignment of Unimodal Medical Text and Vision Representations
Mar 06, 2025Abstract:General-purpose AI models, particularly those designed for text and vision, demonstrate impressive versatility across a wide range of deep-learning tasks. However, they often underperform in specialised domains like medical imaging, where domain-specific solutions or alternative knowledge transfer approaches are typically required. Recent studies have noted that general-purpose models can exhibit similar latent spaces when processing semantically related data, although this alignment does not occur naturally. Building on this insight, it has been shown that applying a simple transformation - at most affine - estimated from a subset of semantically corresponding samples, known as anchors, enables model stitching across diverse training paradigms, architectures, and modalities. In this paper, we explore how semantic alignment - estimating transformations between anchors - can bridge general-purpose AI with specialised medical knowledge. Using multiple public chest X-ray datasets, we demonstrate that model stitching across model architectures allows general models to integrate domain-specific knowledge without additional training, leading to improved performance on medical tasks. Furthermore, we introduce a novel zero-shot classification approach for unimodal vision encoders that leverages semantic alignment across modalities. Our results show that our method not only outperforms general multimodal models but also approaches the performance levels of fully trained, medical-specific multimodal solutions
High-dimensional multimodal uncertainty estimation by manifold alignment:Application to 3D right ventricular strain computations
Jan 21, 2025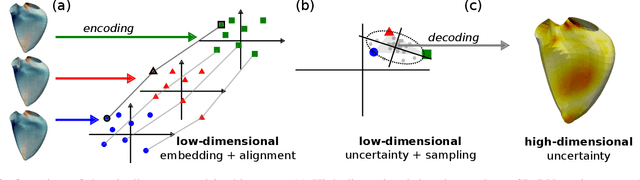
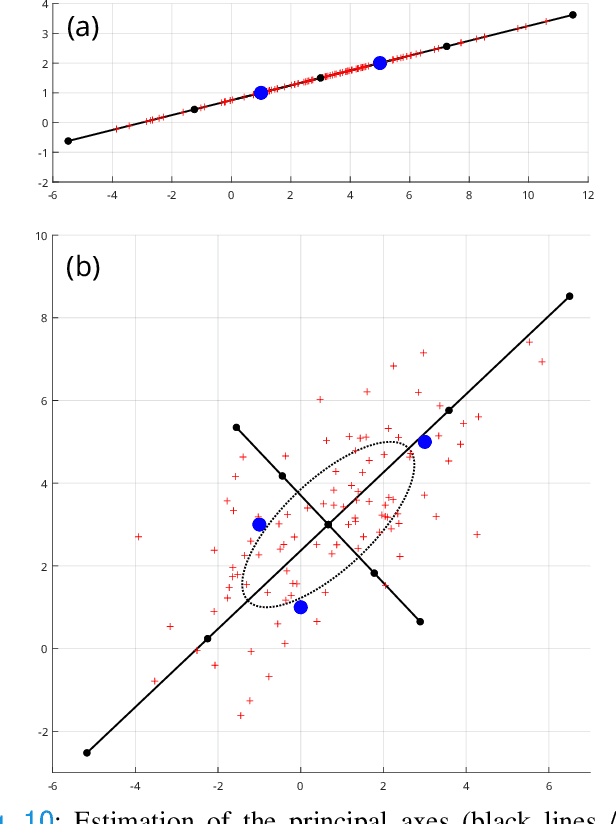
Abstract:Confidence in the results is a key ingredient to improve the adoption of machine learning methods by clinicians. Uncertainties on the results have been considered in the literature, but mostly those originating from the learning and processing methods. Uncertainty on the data is hardly challenged, as a single sample is often considered representative enough of each subject included in the analysis. In this paper, we propose a representation learning strategy to estimate local uncertainties on a physiological descriptor (here, myocardial deformation) previously obtained from medical images by different definitions or computations. We first use manifold alignment to match the latent representations associated to different high-dimensional input descriptors. Then, we formulate plausible distributions of latent uncertainties, and finally exploit them to reconstruct uncertainties on the input high-dimensional descriptors. We demonstrate its relevance for the quantification of myocardial deformation (strain) from 3D echocardiographic image sequences of the right ventricle, for which a lack of consensus exists in its definition and which directional component to use. We used a database of 100 control subjects with right ventricle overload, for which different types of strain are available at each point of the right ventricle endocardial surface mesh. Our approach quantifies local uncertainties on myocardial deformation from different descriptors defining this physiological concept. Such uncertainties cannot be directly estimated by local statistics on such descriptors, potentially of heterogeneous types. Beyond this controlled illustrative application, our methodology has the potential to be generalized to many other population analyses considering heterogeneous high-dimensional descriptors.
On Differentially Private 3D Medical Image Synthesis with Controllable Latent Diffusion Models
Jul 23, 2024
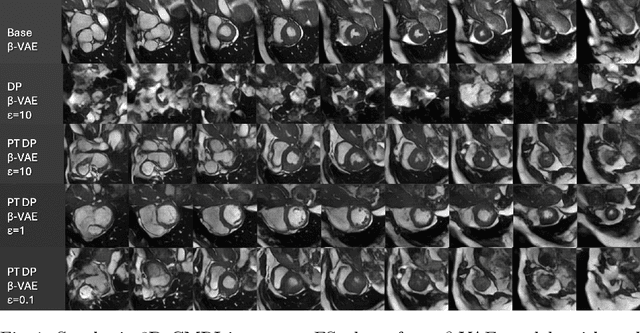
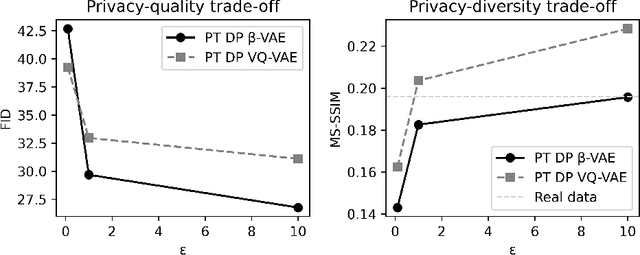

Abstract:Generally, the small size of public medical imaging datasets coupled with stringent privacy concerns, hampers the advancement of data-hungry deep learning models in medical imaging. This study addresses these challenges for 3D cardiac MRI images in the short-axis view. We propose Latent Diffusion Models that generate synthetic images conditioned on medical attributes, while ensuring patient privacy through differentially private model training. To our knowledge, this is the first work to apply and quantify differential privacy in 3D medical image generation. We pre-train our models on public data and finetune them with differential privacy on the UK Biobank dataset. Our experiments reveal that pre-training significantly improves model performance, achieving a Fr\'echet Inception Distance (FID) of 26.77 at $\epsilon=10$, compared to 92.52 for models without pre-training. Additionally, we explore the trade-off between privacy constraints and image quality, investigating how tighter privacy budgets affect output controllability and may lead to degraded performance. Our results demonstrate that proper consideration during training with differential privacy can substantially improve the quality of synthetic cardiac MRI images, but there are still notable challenges in achieving consistent medical realism.
Interpretable Representation Learning of Cardiac MRI via Attribute Regularization
Jun 12, 2024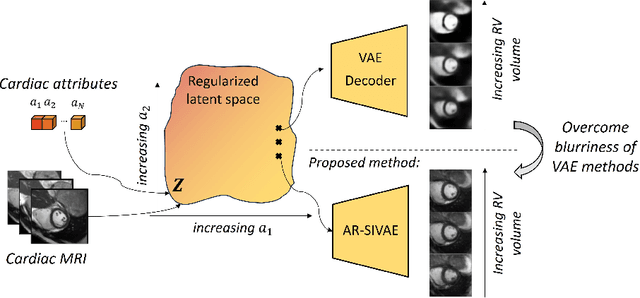
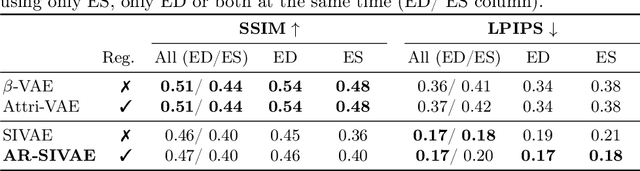
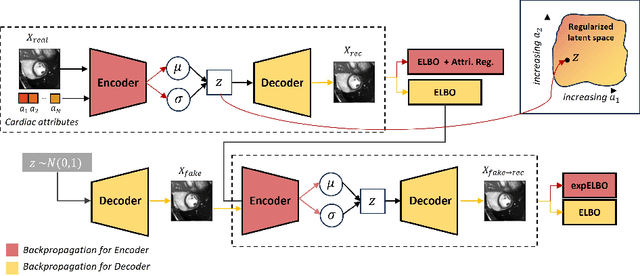
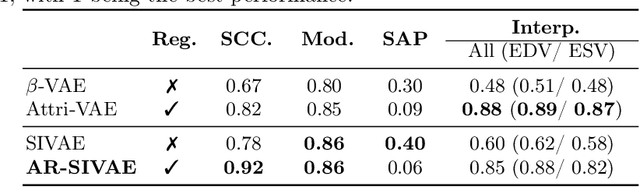
Abstract:Interpretability is essential in medical imaging to ensure that clinicians can comprehend and trust artificial intelligence models. Several approaches have been recently considered to encode attributes in the latent space to enhance its interpretability. Notably, attribute regularization aims to encode a set of attributes along the dimensions of a latent representation. However, this approach is based on Variational AutoEncoder and suffers from blurry reconstruction. In this paper, we propose an Attributed-regularized Soft Introspective Variational Autoencoder that combines attribute regularization of the latent space within the framework of an adversarially trained variational autoencoder. We demonstrate on short-axis cardiac Magnetic Resonance images of the UK Biobank the ability of the proposed method to address blurry reconstruction issues of variational autoencoder methods while preserving the latent space interpretability.
Influence of Prompting Strategies on Segment Anything Model (SAM) for Short-axis Cardiac MRI segmentation
Dec 14, 2023Abstract:The Segment Anything Model (SAM) has recently emerged as a significant breakthrough in foundation models, demonstrating remarkable zero-shot performance in object segmentation tasks. While SAM is designed for generalization, it exhibits limitations in handling specific medical imaging tasks that require fine-structure segmentation or precise boundaries. In this paper, we focus on the task of cardiac magnetic resonance imaging (cMRI) short-axis view segmentation using the SAM foundation model. We conduct a comprehensive investigation of the impact of different prompting strategies (including bounding boxes, positive points, negative points, and their combinations) on segmentation performance. We evaluate on two public datasets using the baseline model and models fine-tuned with varying amounts of annotated data, ranging from a limited number of volumes to a fully annotated dataset. Our findings indicate that prompting strategies significantly influence segmentation performance. Combining positive points with either bounding boxes or negative points shows substantial benefits, but little to no benefit when combined simultaneously. We further observe that fine-tuning SAM with a few annotated volumes improves segmentation performance when properly prompted. Specifically, fine-tuning with bounding boxes has a positive impact, while fine-tuning without bounding boxes leads to worse results compared to baseline.
Attribute Regularized Soft Introspective Variational Autoencoder for Interpretable Cardiac Disease Classification
Dec 14, 2023Abstract:Interpretability is essential in medical imaging to ensure that clinicians can comprehend and trust artificial intelligence models. In this paper, we propose a novel interpretable approach that combines attribute regularization of the latent space within the framework of an adversarially trained variational autoencoder. Comparative experiments on a cardiac MRI dataset demonstrate the ability of the proposed method to address blurry reconstruction issues of variational autoencoder methods and improve latent space interpretability. Additionally, our analysis of a downstream task reveals that the classification of cardiac disease using the regularized latent space heavily relies on attribute regularized dimensions, demonstrating great interpretability by connecting the used attributes for prediction with clinical observations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge