Klaus Kofoed
NUDF: Neural Unsigned Distance Fields for high resolution 3D medical image segmentation
Apr 25, 2025Abstract:Medical image segmentation is often considered as the task of labelling each pixel or voxel as being inside or outside a given anatomy. Processing the images at their original size and resolution often result in insuperable memory requirements, but downsampling the images leads to a loss of important details. Instead of aiming to represent a smooth and continuous surface in a binary voxel-grid, we propose to learn a Neural Unsigned Distance Field (NUDF) directly from the image. The small memory requirements of NUDF allow for high resolution processing, while the continuous nature of the distance field allows us to create high resolution 3D mesh models of shapes of any topology (i.e. open surfaces). We evaluate our method on the task of left atrial appendage (LAA) segmentation from Computed Tomography (CT) images. The LAA is a complex and highly variable shape, being thus difficult to represent with traditional segmentation methods using discrete labelmaps. With our proposed method, we are able to predict 3D mesh models that capture the details of the LAA and achieve accuracy in the order of the voxel spacing in the CT images.
Spatio-temporal neural distance fields for conditional generative modeling of the heart
Jul 15, 2024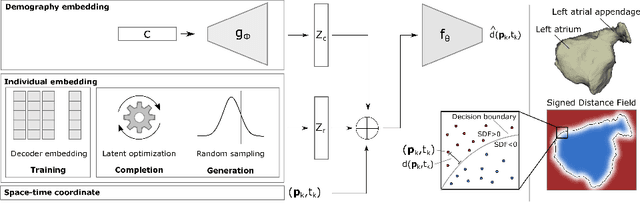
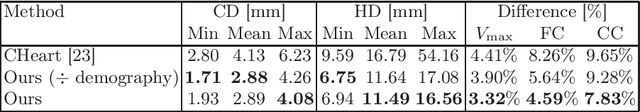
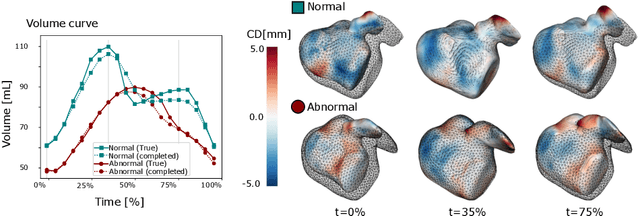
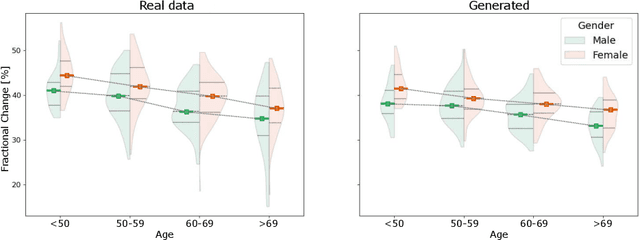
Abstract:The rhythmic pumping motion of the heart stands as a cornerstone in life, as it circulates blood to the entire human body through a series of carefully timed contractions of the individual chambers. Changes in the size, shape and movement of the chambers can be important markers for cardiac disease and modeling this in relation to clinical demography or disease is therefore of interest. Existing methods for spatio-temporal modeling of the human heart require shape correspondence over time or suffer from large memory requirements, making it difficult to use for complex anatomies. We introduce a novel conditional generative model, where the shape and movement is modeled implicitly in the form of a spatio-temporal neural distance field and conditioned on clinical demography. The model is based on an auto-decoder architecture and aims to disentangle the individual variations from that related to the clinical demography. It is tested on the left atrium (including the left atrial appendage), where it outperforms current state-of-the-art methods for anatomical sequence completion and generates synthetic sequences that realistically mimics the shape and motion of the real left atrium. In practice, this means we can infer functional measurements from a static image, generate synthetic populations with specified demography or disease and investigate how non-imaging clinical data effect the shape and motion of cardiac anatomies.
Signed Distance Field based Segmentation and Statistical Shape Modelling of the Left Atrial Appendage
Feb 12, 2024Abstract:Patients with atrial fibrillation have a 5-7 fold increased risk of having an ischemic stroke. In these cases, the most common site of thrombus localization is inside the left atrial appendage (LAA) and studies have shown a correlation between the LAA shape and the risk of ischemic stroke. These studies make use of manual measurement and qualitative assessment of shape and are therefore prone to large inter-observer discrepancies, which may explain the contradictions between the conclusions in different studies. We argue that quantitative shape descriptors are necessary to robustly characterize LAA morphology and relate to other functional parameters and stroke risk. Deep Learning methods are becoming standardly available for segmenting cardiovascular structures from high resolution images such as computed tomography (CT), but only few have been tested for LAA segmentation. Furthermore, the majority of segmentation algorithms produces non-smooth 3D models that are not ideal for further processing, such as statistical shape analysis or computational fluid modelling. In this paper we present a fully automatic pipeline for image segmentation, mesh model creation and statistical shape modelling of the LAA. The LAA anatomy is implicitly represented as a signed distance field (SDF), which is directly regressed from the CT image using Deep Learning. The SDF is further used for registering the LAA shapes to a common template and build a statistical shape model (SSM). Based on 106 automatically segmented LAAs, the built SSM reveals that the LAA shape can be quantified using approximately 5 PCA modes and allows the identification of two distinct shape clusters corresponding to the so-called chicken-wing and non-chicken-wing morphologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge