Julia Machnio
Deep Learning-Based Regional White Matter Hyperintensity Mapping as a Robust Biomarker for Alzheimer's Disease
Nov 18, 2025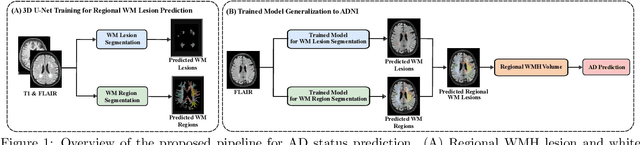
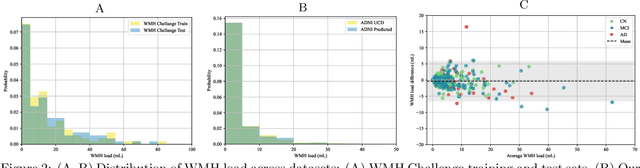
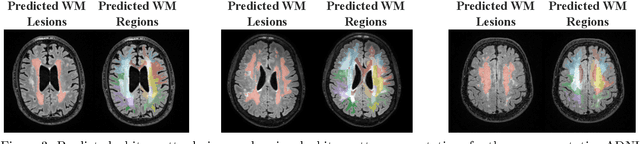
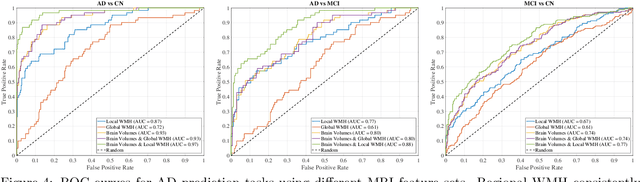
Abstract:White matter hyperintensities (WMH) are key imaging markers in cognitive aging, Alzheimer's disease (AD), and related dementias. Although automated methods for WMH segmentation have advanced, most provide only global lesion load and overlook their spatial distribution across distinct white matter regions. We propose a deep learning framework for robust WMH segmentation and localization, evaluated across public datasets and an independent Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort. Our results show that the predicted lesion loads are in line with the reference WMH estimates, confirming the robustness to variations in lesion load, acquisition, and demographics. Beyond accurate segmentation, we quantify WMH load within anatomically defined regions and combine these measures with brain structure volumes to assess diagnostic value. Regional WMH volumes consistently outperform global lesion burden for disease classification, and integration with brain atrophy metrics further improves performance, reaching area under the curve (AUC) values up to 0.97. Several spatially distinct regions, particularly within anterior white matter tracts, are reproducibly associated with diagnostic status, indicating localized vulnerability in AD. These results highlight the added value of regional WMH quantification. Incorporating localized lesion metrics alongside atrophy markers may enhance early diagnosis and stratification in neurodegenerative disorders.
MRI Embeddings Complement Clinical Predictors for Cognitive Decline Modeling in Alzheimer's Disease Cohorts
Nov 18, 2025Abstract:Accurate modeling of cognitive decline in Alzheimer's disease is essential for early stratification and personalized management. While tabular predictors provide robust markers of global risk, their ability to capture subtle brain changes remains limited. In this study, we evaluate the predictive contributions of tabular and imaging-based representations, with a focus on transformer-derived Magnetic Resonance Imaging (MRI) embeddings. We introduce a trajectory-aware labeling strategy based on Dynamic Time Warping clustering to capture heterogeneous patterns of cognitive change, and train a 3D Vision Transformer (ViT) via unsupervised reconstruction on harmonized and augmented MRI data to obtain anatomy-preserving embeddings without progression labels. The pretrained encoder embeddings are subsequently assessed using both traditional machine learning classifiers and deep learning heads, and compared against tabular representations and convolutional network baselines. Results highlight complementary strengths across modalities. Clinical and volumetric features achieved the highest AUCs of around 0.70 for predicting mild and severe progression, underscoring their utility in capturing global decline trajectories. In contrast, MRI embeddings from the ViT model were most effective in distinguishing cognitively stable individuals with an AUC of 0.71. However, all approaches struggled in the heterogeneous moderate group. These findings indicate that clinical features excel in identifying high-risk extremes, whereas transformer-based MRI embeddings are more sensitive to subtle markers of stability, motivating multimodal fusion strategies for AD progression modeling.
A large-scale heterogeneous 3D magnetic resonance brain imaging dataset for self-supervised learning
Jun 17, 2025Abstract:We present FOMO60K, a large-scale, heterogeneous dataset of 60,529 brain Magnetic Resonance Imaging (MRI) scans from 13,900 sessions and 11,187 subjects, aggregated from 16 publicly available sources. The dataset includes both clinical- and research-grade images, multiple MRI sequences, and a wide range of anatomical and pathological variability, including scans with large brain anomalies. Minimal preprocessing was applied to preserve the original image characteristics while reducing barriers to entry for new users. Accompanying code for self-supervised pretraining and finetuning is provided. FOMO60K is intended to support the development and benchmarking of self-supervised learning methods in medical imaging at scale.
MICCAI-CDMRI 2023 QuantConn Challenge Findings on Achieving Robust Quantitative Connectivity through Harmonized Preprocessing of Diffusion MRI
Nov 14, 2024



Abstract:White matter alterations are increasingly implicated in neurological diseases and their progression. International-scale studies use diffusion-weighted magnetic resonance imaging (DW-MRI) to qualitatively identify changes in white matter microstructure and connectivity. Yet, quantitative analysis of DW-MRI data is hindered by inconsistencies stemming from varying acquisition protocols. There is a pressing need to harmonize the preprocessing of DW-MRI datasets to ensure the derivation of robust quantitative diffusion metrics across acquisitions. In the MICCAI-CDMRI 2023 QuantConn challenge, participants were provided raw data from the same individuals collected on the same scanner but with two different acquisitions and tasked with preprocessing the DW-MRI to minimize acquisition differences while retaining biological variation. Submissions are evaluated on the reproducibility and comparability of cross-acquisition bundle-wise microstructure measures, bundle shape features, and connectomics. The key innovations of the QuantConn challenge are that (1) we assess bundles and tractography in the context of harmonization for the first time, (2) we assess connectomics in the context of harmonization for the first time, and (3) we have 10x additional subjects over prior harmonization challenge, MUSHAC and 100x over SuperMUDI. We find that bundle surface area, fractional anisotropy, connectome assortativity, betweenness centrality, edge count, modularity, nodal strength, and participation coefficient measures are most biased by acquisition and that machine learning voxel-wise correction, RISH mapping, and NeSH methods effectively reduce these biases. In addition, microstructure measures AD, MD, RD, bundle length, connectome density, efficiency, and path length are least biased by these acquisition differences.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024/019
Yucca: A Deep Learning Framework For Medical Image Analysis
Jul 29, 2024


Abstract:Medical image analysis using deep learning frameworks has advanced healthcare by automating complex tasks, but many existing frameworks lack flexibility, modularity, and user-friendliness. To address these challenges, we introduce Yucca, an open-source AI framework available at https://github.com/Sllambias/yucca, designed specifically for medical imaging applications and built on PyTorch and PyTorch Lightning. Yucca features a three-tiered architecture: Functional, Modules, and Pipeline, providing a comprehensive and customizable solution. Evaluated across diverse tasks such as cerebral microbleeds detection, white matter hyperintensity segmentation, and hippocampus segmentation, Yucca achieves state-of-the-art results, demonstrating its robustness and versatility. Yucca offers a powerful, flexible, and user-friendly platform for medical image analysis, inviting community contributions to advance its capabilities and impact.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge