Thomas C Booth
Reinforcement Learning for Safe Autonomous Two Device Navigation of Cerebral Vessels in Mechanical Thrombectomy
Mar 31, 2025Abstract:Purpose: Autonomous systems in mechanical thrombectomy (MT) hold promise for reducing procedure times, minimizing radiation exposure, and enhancing patient safety. However, current reinforcement learning (RL) methods only reach the carotid arteries, are not generalizable to other patient vasculatures, and do not consider safety. We propose a safe dual-device RL algorithm that can navigate beyond the carotid arteries to cerebral vessels. Methods: We used the Simulation Open Framework Architecture to represent the intricacies of cerebral vessels, and a modified Soft Actor-Critic RL algorithm to learn, for the first time, the navigation of micro-catheters and micro-guidewires. We incorporate patient safety metrics into our reward function by integrating guidewire tip forces. Inverse RL is used with demonstrator data on 12 patient-specific vascular cases. Results: Our simulation demonstrates successful autonomous navigation within unseen cerebral vessels, achieving a 96% success rate, 7.0s procedure time, and 0.24 N mean forces, well below the proposed 1.5 N vessel rupture threshold. Conclusion: To the best of our knowledge, our proposed autonomous system for MT two-device navigation reaches cerebral vessels, considers safety, and is generalizable to unseen patient-specific cases for the first time. We envisage future work will extend the validation to vasculatures of different complexity and on in vitro models. While our contributions pave the way towards deploying agents in clinical settings, safety and trustworthiness will be crucial elements to consider when proposing new methodology.
Machine learning algorithms to predict the risk of rupture of intracranial aneurysms: a systematic review
Dec 06, 2024Abstract:Purpose: Subarachnoid haemorrhage is a potentially fatal consequence of intracranial aneurysm rupture, however, it is difficult to predict if aneurysms will rupture. Prophylactic treatment of an intracranial aneurysm also involves risk, hence identifying rupture-prone aneurysms is of substantial clinical importance. This systematic review aims to evaluate the performance of machine learning algorithms for predicting intracranial aneurysm rupture risk. Methods: MEDLINE, Embase, Cochrane Library and Web of Science were searched until December 2023. Studies incorporating any machine learning algorithm to predict the risk of rupture of an intracranial aneurysm were included. Risk of bias was assessed using the Prediction Model Risk of Bias Assessment Tool (PROBAST). PROSPERO registration: CRD42023452509. Results: Out of 10,307 records screened, 20 studies met the eligibility criteria for this review incorporating a total of 20,286 aneurysm cases. The machine learning models gave a 0.66-0.90 range for performance accuracy. The models were compared to current clinical standards in six studies and gave mixed results. Most studies posed high or unclear risks of bias and concerns for applicability, limiting the inferences that can be drawn from them. There was insufficient homogenous data for a meta-analysis. Conclusions: Machine learning can be applied to predict the risk of rupture for intracranial aneurysms. However, the evidence does not comprehensively demonstrate superiority to existing practice, limiting its role as a clinical adjunct. Further prospective multicentre studies of recent machine learning tools are needed to prove clinical validation before they are implemented in the clinic.
Artificial intelligence for abnormality detection in high volume neuroimaging: a systematic review and meta-analysis
May 09, 2024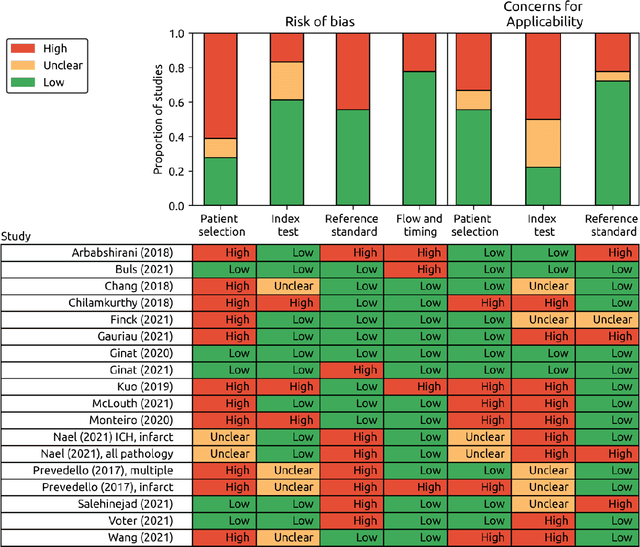
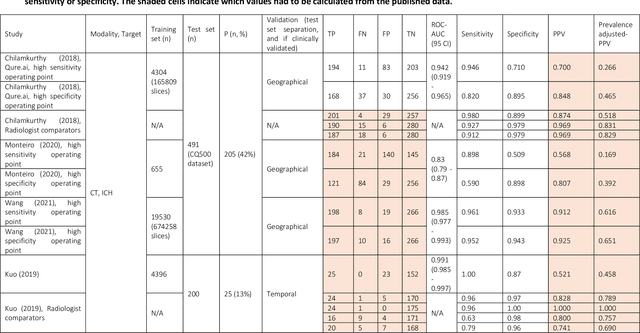
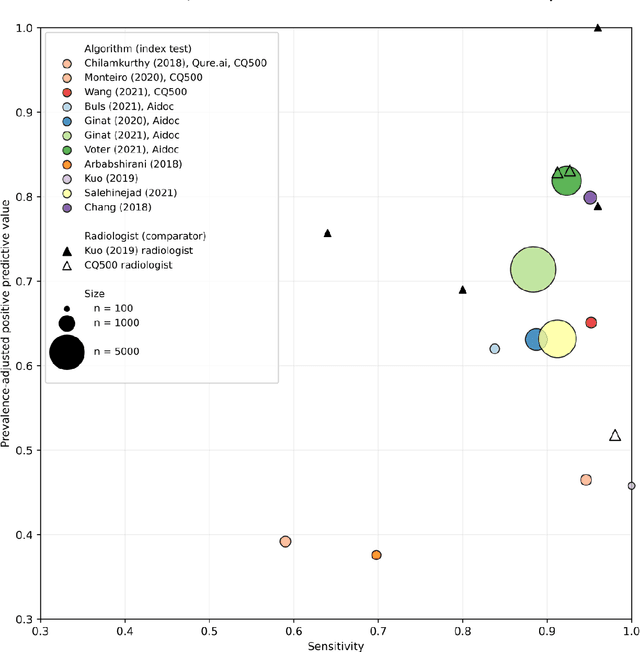
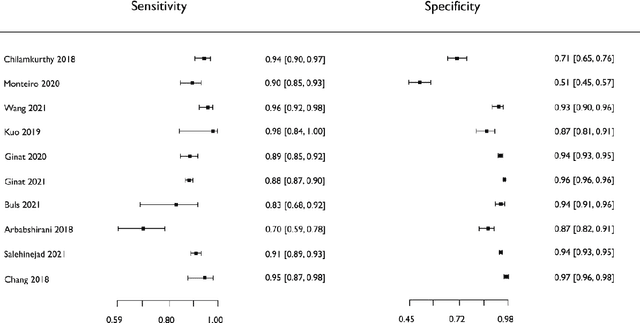
Abstract:Purpose: Most studies evaluating artificial intelligence (AI) models that detect abnormalities in neuroimaging are either tested on unrepresentative patient cohorts or are insufficiently well-validated, leading to poor generalisability to real-world tasks. The aim was to determine the diagnostic test accuracy and summarise the evidence supporting the use of AI models performing first-line, high-volume neuroimaging tasks. Methods: Medline, Embase, Cochrane library and Web of Science were searched until September 2021 for studies that temporally or externally validated AI capable of detecting abnormalities in first-line CT or MR neuroimaging. A bivariate random-effects model was used for meta-analysis where appropriate. PROSPERO: CRD42021269563. Results: Only 16 studies were eligible for inclusion. Included studies were not compromised by unrepresentative datasets or inadequate validation methodology. Direct comparison with radiologists was available in 4/16 studies. 15/16 had a high risk of bias. Meta-analysis was only suitable for intracranial haemorrhage detection in CT imaging (10/16 studies), where AI systems had a pooled sensitivity and specificity 0.90 (95% CI 0.85 - 0.94) and 0.90 (95% CI 0.83 - 0.95) respectively. Other AI studies using CT and MRI detected target conditions other than haemorrhage (2/16), or multiple target conditions (4/16). Only 3/16 studies implemented AI in clinical pathways, either for pre-read triage or as post-read discrepancy identifiers. Conclusion: The paucity of eligible studies reflects that most abnormality detection AI studies were not adequately validated in representative clinical cohorts. The few studies describing how abnormality detection AI could impact patients and clinicians did not explore the full ramifications of clinical implementation.
Letter to the Editor: What are the legal and ethical considerations of submitting radiology reports to ChatGPT?
May 09, 2024Abstract:This letter critically examines the recent article by Infante et al. assessing the utility of large language models (LLMs) like GPT-4, Perplexity, and Bard in identifying urgent findings in emergency radiology reports. While acknowledging the potential of LLMs in generating labels for computer vision, concerns are raised about the ethical implications of using patient data without explicit approval, highlighting the necessity of stringent data protection measures under GDPR.
Overcoming challenges of translating deep-learning models for glioblastoma: the ZGBM consortium
May 07, 2024Abstract:Objective: To report imaging protocol and scheduling variance in routine care of glioblastoma patients in order to demonstrate challenges of integrating deep-learning models in glioblastoma care pathways. Additionally, to understand the most common imaging studies and image contrasts to inform the development of potentially robust deep-learning models. Methods: MR imaging data were analysed from a random sample of five patients from the prospective cohort across five participating sites of the ZGBM consortium. Reported clinical and treatment data alongside DICOM header information were analysed to understand treatment pathway imaging schedules. Results: All sites perform all structural imaging at every stage in the pathway except for the presurgical study, where in some sites only contrast-enhanced T1-weighted imaging is performed. Diffusion MRI is the most common non-structural imaging type, performed at every site. Conclusion: The imaging protocol and scheduling varies across the UK, making it challenging to develop machine-learning models that could perform robustly at other centres. Structural imaging is performed most consistently across all centres. Advances in knowledge: Successful translation of deep-learning models will likely be based on structural post-treatment imaging unless there is significant effort made to standardise non-structural or peri-operative imaging protocols and schedules.
Artificial Intelligence in the Autonomous Navigation of Endovascular Interventions: A Systematic Review
May 06, 2024



Abstract:Purpose: Autonomous navigation of devices in endovascular interventions can decrease operation times, improve decision-making during surgery, and reduce operator radiation exposure while increasing access to treatment. This systematic review explores recent literature to assess the impact, challenges, and opportunities artificial intelligence (AI) has for the autonomous endovascular intervention navigation. Methods: PubMed and IEEEXplore databases were queried. Eligibility criteria included studies investigating the use of AI in enabling the autonomous navigation of catheters/guidewires in endovascular interventions. Following PRISMA, articles were assessed using QUADAS-2. PROSPERO: CRD42023392259. Results: Among 462 studies, fourteen met inclusion criteria. Reinforcement learning (9/14, 64%) and learning from demonstration (7/14, 50%) were used as data-driven models for autonomous navigation. Studies predominantly utilised physical phantoms (10/14, 71%) and in silico (4/14, 29%) models. Experiments within or around the blood vessels of the heart were reported by the majority of studies (10/14, 71%), while simple non-anatomical vessel platforms were used in three studies (3/14, 21%), and the porcine liver venous system in one study. We observed that risk of bias and poor generalisability were present across studies. No procedures were performed on patients in any of the studies reviewed. Studies lacked patient selection criteria, reference standards, and reproducibility, resulting in low clinical evidence levels. Conclusions: AI's potential in autonomous endovascular navigation is promising, but in an experimental proof-of-concept stage, with a technology readiness level of 3. We highlight that reference standards with well-identified performance metrics are crucial to allow for comparisons of data-driven algorithms proposed in the years to come.
* Abstract shortened for arXiv character limit
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge