Alysha Chelliah
Overcoming challenges of translating deep-learning models for glioblastoma: the ZGBM consortium
May 07, 2024Abstract:Objective: To report imaging protocol and scheduling variance in routine care of glioblastoma patients in order to demonstrate challenges of integrating deep-learning models in glioblastoma care pathways. Additionally, to understand the most common imaging studies and image contrasts to inform the development of potentially robust deep-learning models. Methods: MR imaging data were analysed from a random sample of five patients from the prospective cohort across five participating sites of the ZGBM consortium. Reported clinical and treatment data alongside DICOM header information were analysed to understand treatment pathway imaging schedules. Results: All sites perform all structural imaging at every stage in the pathway except for the presurgical study, where in some sites only contrast-enhanced T1-weighted imaging is performed. Diffusion MRI is the most common non-structural imaging type, performed at every site. Conclusion: The imaging protocol and scheduling varies across the UK, making it challenging to develop machine-learning models that could perform robustly at other centres. Structural imaging is performed most consistently across all centres. Advances in knowledge: Successful translation of deep-learning models will likely be based on structural post-treatment imaging unless there is significant effort made to standardise non-structural or peri-operative imaging protocols and schedules.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
Machine Learning and Glioblastoma: Treatment Response Monitoring Biomarkers in 2021
Apr 15, 2021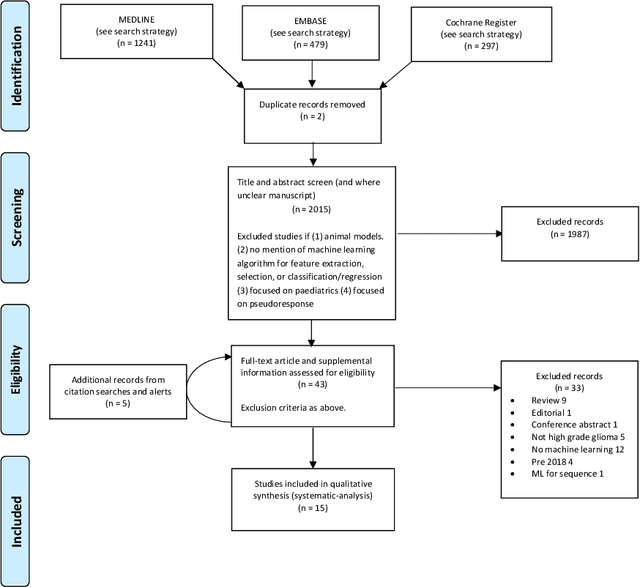
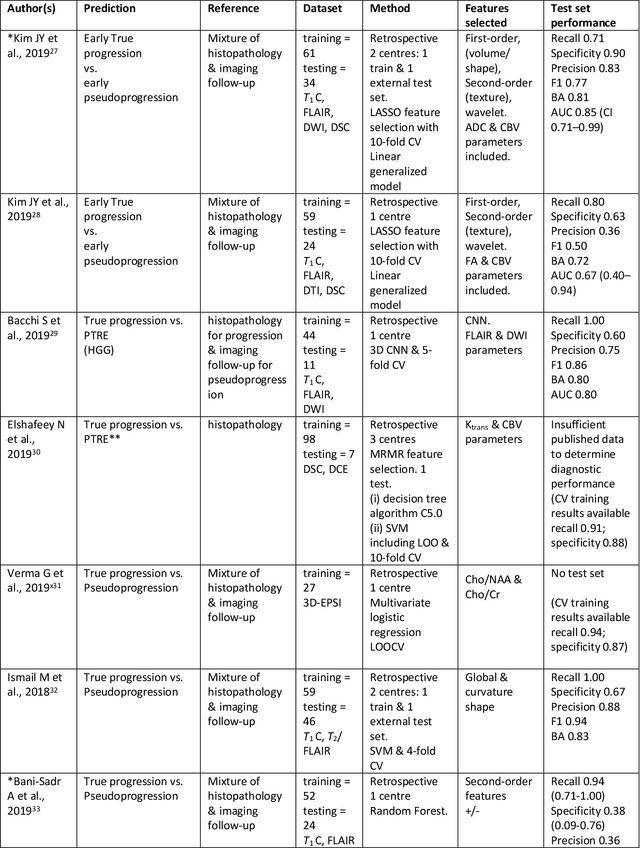
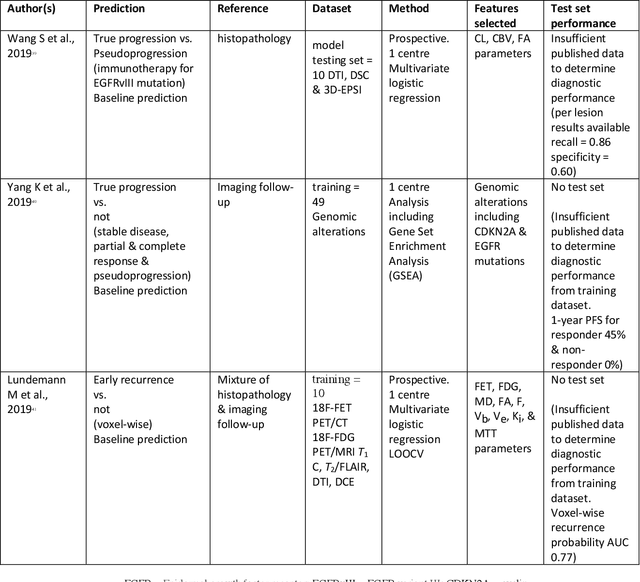
Abstract:The aim of the systematic review was to assess recently published studies on diagnostic test accuracy of glioblastoma treatment response monitoring biomarkers in adults, developed through machine learning (ML). Articles were searched for using MEDLINE, EMBASE, and the Cochrane Register. Included study participants were adult patients with high grade glioma who had undergone standard treatment (maximal resection, radiotherapy with concomitant and adjuvant temozolomide) and subsequently underwent follow-up imaging to determine treatment response status. Risk of bias and applicability was assessed with QUADAS 2 methodology. Contingency tables were created for hold-out test sets and recall, specificity, precision, F1-score, balanced accuracy calculated. Fifteen studies were included with 1038 patients in training sets and 233 in test sets. To determine whether there was progression or a mimic, the reference standard combination of follow-up imaging and histopathology at re-operation was applied in 67% of studies. The small numbers of patient included in studies, the high risk of bias and concerns of applicability in the study designs (particularly in relation to the reference standard and patient selection due to confounding), and the low level of evidence, suggest that limited conclusions can be drawn from the data. There is likely good diagnostic performance of machine learning models that use MRI features to distinguish between progression and mimics. The diagnostic performance of ML using implicit features did not appear to be superior to ML using explicit features. There are a range of ML-based solutions poised to become treatment response monitoring biomarkers for glioblastoma. To achieve this, the development and validation of ML models require large, well-annotated datasets where the potential for confounding in the study design has been carefully considered.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge