James Cole
Disentangled Diffusion Autoencoder for Harmonization of Multi-site Neuroimaging Data
Aug 28, 2024



Abstract:Combining neuroimaging datasets from multiple sites and scanners can help increase statistical power and thus provide greater insight into subtle neuroanatomical effects. However, site-specific effects pose a challenge by potentially obscuring the biological signal and introducing unwanted variance. Existing harmonization techniques, which use statistical models to remove such effects, have been shown to incompletely remove site effects while also failing to preserve biological variability. More recently, generative models using GANs or autoencoder-based approaches, have been proposed for site adjustment. However, such methods are known for instability during training or blurry image generation. In recent years, diffusion models have become increasingly popular for their ability to generate high-quality synthetic images. In this work, we introduce the disentangled diffusion autoencoder (DDAE), a novel diffusion model designed for controlling specific aspects of an image. We apply the DDAE to the task of harmonizing MR images by generating high-quality site-adjusted images that preserve biological variability. We use data from 7 different sites and demonstrate the DDAE's superiority in generating high-resolution, harmonized 2D MR images over previous approaches. As far as we are aware, this work marks the first diffusion-based model for site adjustment of neuroimaging data.
Normative Diffusion Autoencoders: Application to Amyotrophic Lateral Sclerosis
Jul 19, 2024Abstract:Predicting survival in Amyotrophic Lateral Sclerosis (ALS) is a challenging task. Magnetic resonance imaging (MRI) data provide in vivo insight into brain health, but the low prevalence of the condition and resultant data scarcity limit training set sizes for prediction models. Survival models are further hindered by the subtle and often highly localised profile of ALS-related neurodegeneration. Normative models present a solution as they increase statistical power by leveraging large healthy cohorts. Separately, diffusion models excel in capturing the semantics embedded within images including subtle signs of accelerated brain ageing, which may help predict survival in ALS. Here, we combine the benefits of generative and normative modelling by introducing the normative diffusion autoencoder framework. To our knowledge, this is the first use of normative modelling within a diffusion autoencoder, as well as the first application of normative modelling to ALS. Our approach outperforms generative and non-generative normative modelling benchmarks in ALS prognostication, demonstrating enhanced predictive accuracy in the context of ALS survival prediction and normative modelling in general.
Artificial intelligence for abnormality detection in high volume neuroimaging: a systematic review and meta-analysis
May 09, 2024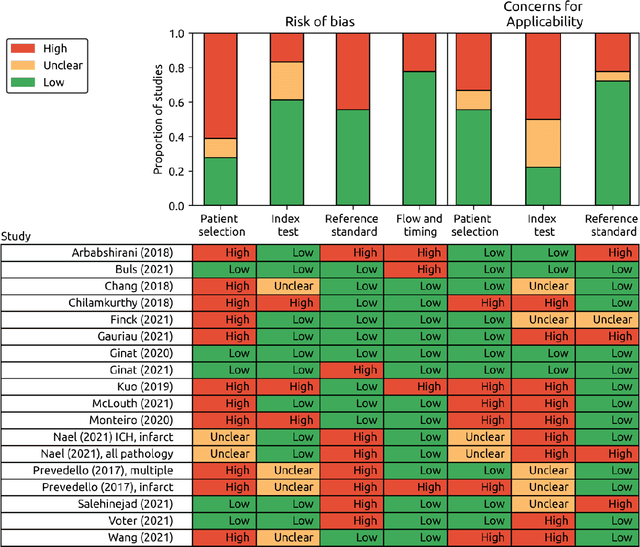
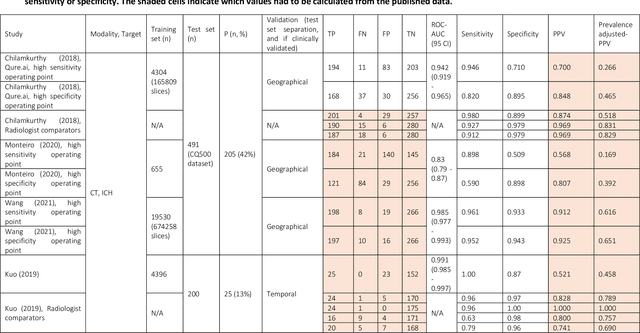
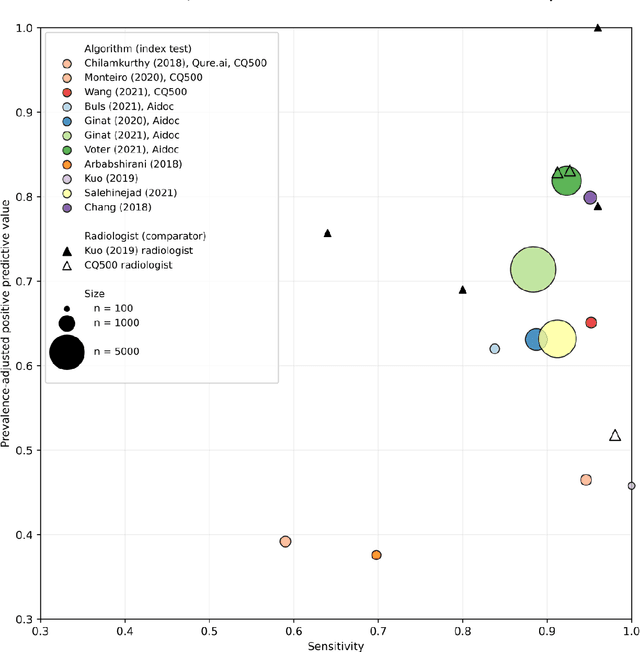
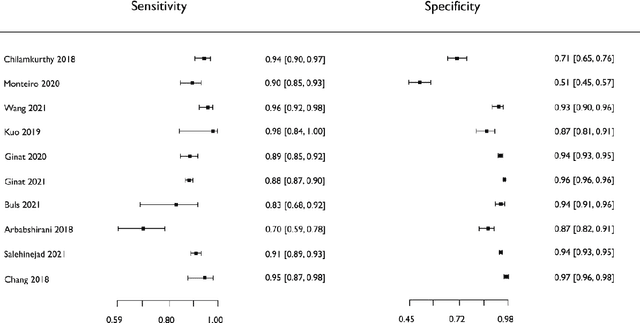
Abstract:Purpose: Most studies evaluating artificial intelligence (AI) models that detect abnormalities in neuroimaging are either tested on unrepresentative patient cohorts or are insufficiently well-validated, leading to poor generalisability to real-world tasks. The aim was to determine the diagnostic test accuracy and summarise the evidence supporting the use of AI models performing first-line, high-volume neuroimaging tasks. Methods: Medline, Embase, Cochrane library and Web of Science were searched until September 2021 for studies that temporally or externally validated AI capable of detecting abnormalities in first-line CT or MR neuroimaging. A bivariate random-effects model was used for meta-analysis where appropriate. PROSPERO: CRD42021269563. Results: Only 16 studies were eligible for inclusion. Included studies were not compromised by unrepresentative datasets or inadequate validation methodology. Direct comparison with radiologists was available in 4/16 studies. 15/16 had a high risk of bias. Meta-analysis was only suitable for intracranial haemorrhage detection in CT imaging (10/16 studies), where AI systems had a pooled sensitivity and specificity 0.90 (95% CI 0.85 - 0.94) and 0.90 (95% CI 0.83 - 0.95) respectively. Other AI studies using CT and MRI detected target conditions other than haemorrhage (2/16), or multiple target conditions (4/16). Only 3/16 studies implemented AI in clinical pathways, either for pre-read triage or as post-read discrepancy identifiers. Conclusion: The paucity of eligible studies reflects that most abnormality detection AI studies were not adequately validated in representative clinical cohorts. The few studies describing how abnormality detection AI could impact patients and clinicians did not explore the full ramifications of clinical implementation.
Semi-Supervised Diffusion Model for Brain Age Prediction
Feb 14, 2024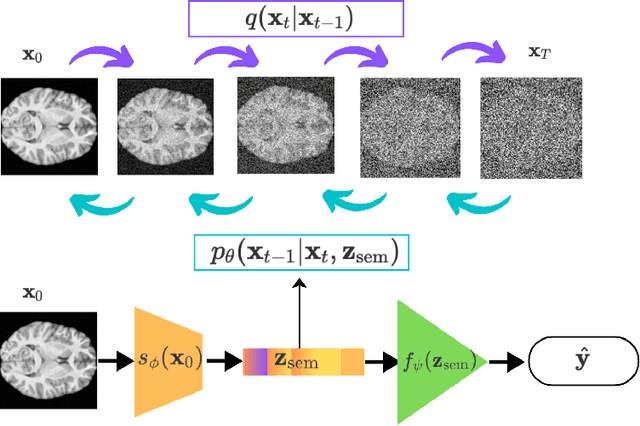

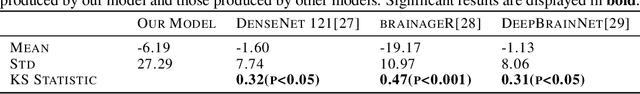
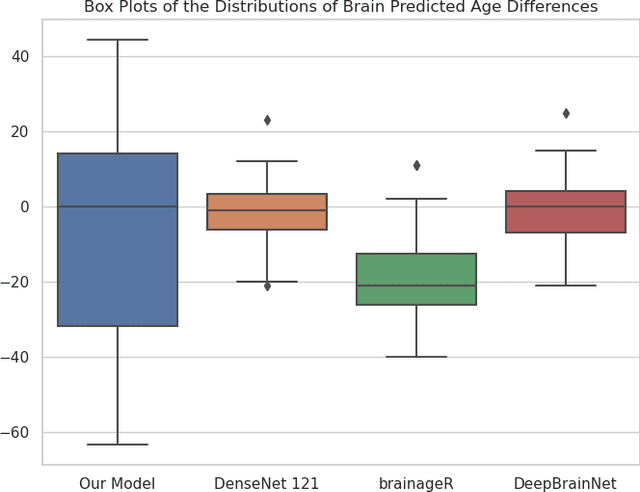
Abstract:Brain age prediction models have succeeded in predicting clinical outcomes in neurodegenerative diseases, but can struggle with tasks involving faster progressing diseases and low quality data. To enhance their performance, we employ a semi-supervised diffusion model, obtaining a 0.83(p<0.01) correlation between chronological and predicted age on low quality T1w MR images. This was competitive with state-of-the-art non-generative methods. Furthermore, the predictions produced by our model were significantly associated with survival length (r=0.24, p<0.05) in Amyotrophic Lateral Sclerosis. Thus, our approach demonstrates the value of diffusion-based architectures for the task of brain age prediction.
Rician likelihood loss for quantitative MRI using self-supervised deep learning
Jul 13, 2023Abstract:Purpose: Previous quantitative MR imaging studies using self-supervised deep learning have reported biased parameter estimates at low SNR. Such systematic errors arise from the choice of Mean Squared Error (MSE) loss function for network training, which is incompatible with Rician-distributed MR magnitude signals. To address this issue, we introduce the negative log Rician likelihood (NLR) loss. Methods: A numerically stable and accurate implementation of the NLR loss was developed to estimate quantitative parameters of the apparent diffusion coefficient (ADC) model and intra-voxel incoherent motion (IVIM) model. Parameter estimation accuracy, precision and overall error were evaluated in terms of bias, variance and root mean squared error and compared against the MSE loss over a range of SNRs (5 - 30). Results: Networks trained with NLR loss show higher estimation accuracy than MSE for the ADC and IVIM diffusion coefficients as SNR decreases, with minimal loss of precision or total error. At high effective SNR (high SNR and small diffusion coefficients), both losses show comparable accuracy and precision for all parameters of both models. Conclusion: The proposed NLR loss is numerically stable and accurate across the full range of tested SNRs and improves parameter estimation accuracy of diffusion coefficients using self-supervised deep learning. We expect the development to benefit quantitative MR imaging techniques broadly, enabling more accurate parameter estimation from noisy data.
Interpretable Alzheimer's Disease Classification Via a Contrastive Diffusion Autoencoder
Jun 05, 2023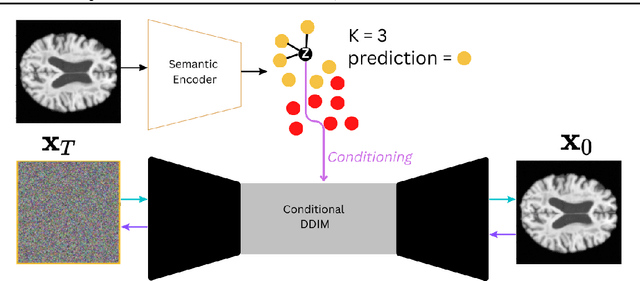
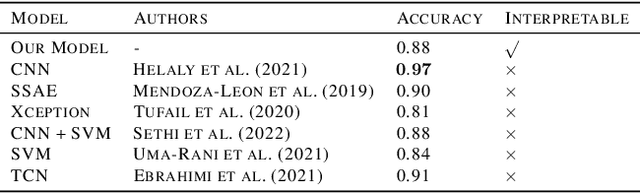
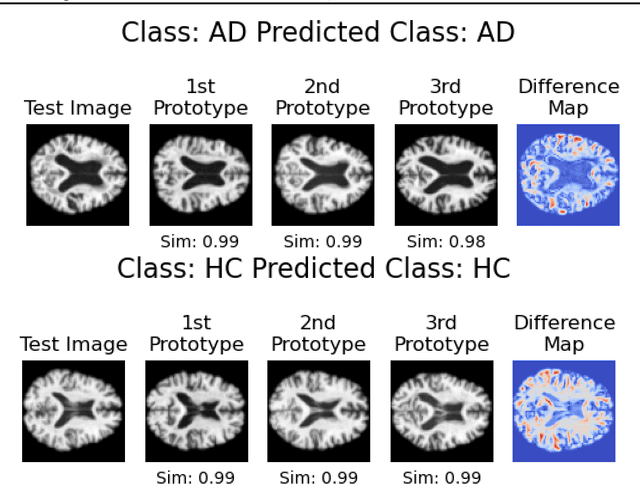
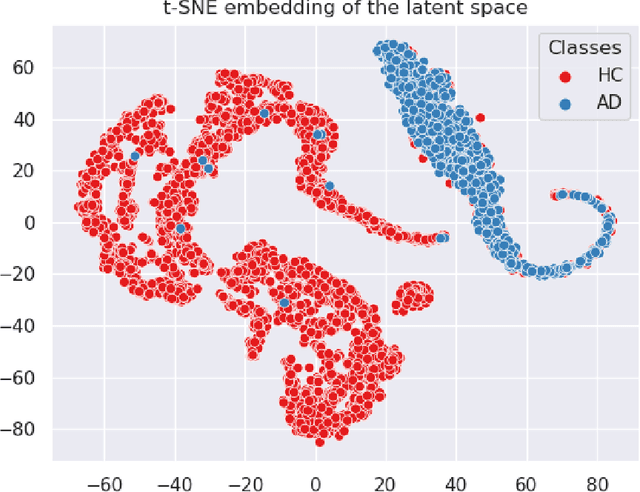
Abstract:In visual object classification, humans often justify their choices by comparing objects to prototypical examples within that class. We may therefore increase the interpretability of deep learning models by imbuing them with a similar style of reasoning. In this work, we apply this principle by classifying Alzheimer's Disease based on the similarity of images to training examples within the latent space. We use a contrastive loss combined with a diffusion autoencoder backbone, to produce a semantically meaningful latent space, such that neighbouring latents have similar image-level features. We achieve a classification accuracy comparable to black box approaches on a dataset of 2D MRI images, whilst producing human interpretable model explanations. Therefore, this work stands as a contribution to the pertinent development of accurate and interpretable deep learning within medical imaging.
Hierarchical Gaussian Processes with Wasserstein-2 Kernels
Oct 28, 2020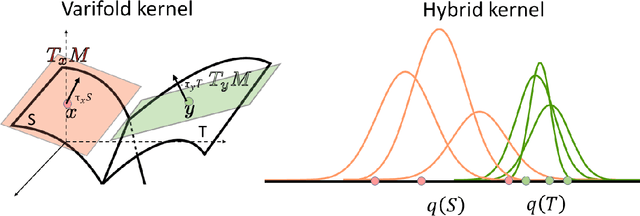
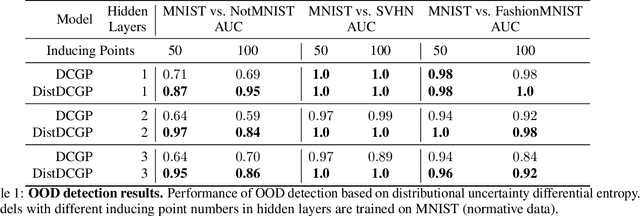
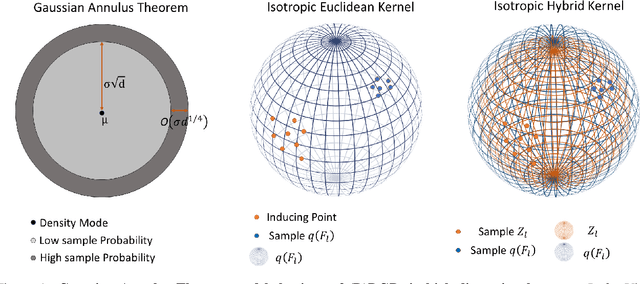
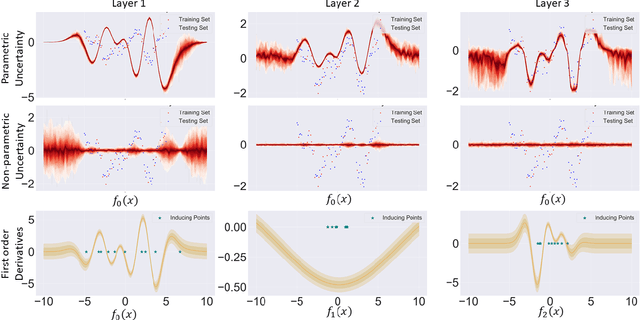
Abstract:We investigate the usefulness of Wasserstein-2 kernels in the context of hierarchical Gaussian Processes. Stemming from an observation that stacking Gaussian Processes severely diminishes the model's ability to detect outliers, which when combined with non-zero mean functions, further extrapolates low variance to regions with low training data density, we posit that directly taking into account the variance in the computation of Wasserstein-2 kernels is of key importance towards maintaining outlier status as we progress through the hierarchy. We propose two new models operating in Wasserstein space which can be seen as equivalents to Deep Kernel Learning and Deep GPs. Through extensive experiments, we show improved performance on large scale datasets and improved out-of-distribution detection on both toy and real data.
NEURO-DRAM: a 3D recurrent visual attention model for interpretable neuroimaging classification
Oct 18, 2019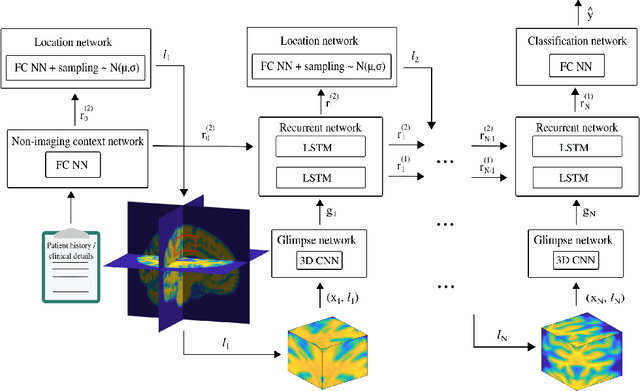
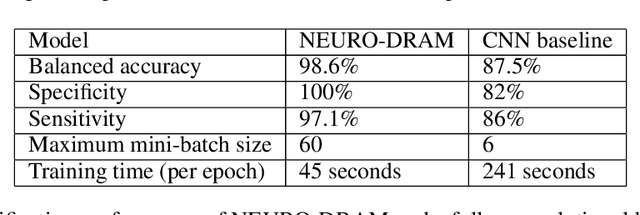

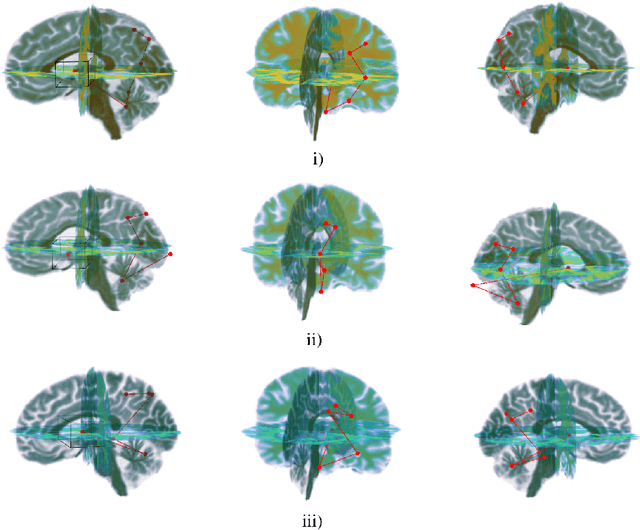
Abstract:Deep learning is attracting significant interest in the neuroimaging community as a means to diagnose psychiatric and neurological disorders from structural magnetic resonance images. However, there is a tendency amongst researchers to adopt architectures optimized for traditional computer vision tasks, rather than design networks customized for neuroimaging data. We address this by introducing NEURO-DRAM, a 3D recurrent visual attention model tailored for neuroimaging classification. The model comprises an agent which, trained by reinforcement learning, learns to navigate through volumetric images, selectively attending to the most informative regions for a given task. When applied to Alzheimer's disease prediction, NEURODRAM achieves state-of-the-art classification accuracy on an out-of-sample dataset, significantly outperforming a baseline convolutional neural network. When further applied to the task of predicting which patients with mild cognitive impairment will be diagnosed with Alzheimer's disease within two years, the model achieves state-of-the-art accuracy with no additional training. Encouragingly, the agent learns, without explicit instruction, a search policy in agreement with standardized radiological hallmarks of Alzheimer's disease, suggesting a route to automated biomarker discovery for more poorly understood disorders.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge