Ayodeji Ijishakin
An X-Ray Is Worth 15 Features: Sparse Autoencoders for Interpretable Radiology Report Generation
Oct 04, 2024Abstract:Radiological services are experiencing unprecedented demand, leading to increased interest in automating radiology report generation. Existing Vision-Language Models (VLMs) suffer from hallucinations, lack interpretability, and require expensive fine-tuning. We introduce SAE-Rad, which uses sparse autoencoders (SAEs) to decompose latent representations from a pre-trained vision transformer into human-interpretable features. Our hybrid architecture combines state-of-the-art SAE advancements, achieving accurate latent reconstructions while maintaining sparsity. Using an off-the-shelf language model, we distil ground-truth reports into radiological descriptions for each SAE feature, which we then compile into a full report for each image, eliminating the need for fine-tuning large models for this task. To the best of our knowledge, SAE-Rad represents the first instance of using mechanistic interpretability techniques explicitly for a downstream multi-modal reasoning task. On the MIMIC-CXR dataset, SAE-Rad achieves competitive radiology-specific metrics compared to state-of-the-art models while using significantly fewer computational resources for training. Qualitative analysis reveals that SAE-Rad learns meaningful visual concepts and generates reports aligning closely with expert interpretations. Our results suggest that SAEs can enhance multimodal reasoning in healthcare, providing a more interpretable alternative to existing VLMs.
Disentangled Diffusion Autoencoder for Harmonization of Multi-site Neuroimaging Data
Aug 28, 2024



Abstract:Combining neuroimaging datasets from multiple sites and scanners can help increase statistical power and thus provide greater insight into subtle neuroanatomical effects. However, site-specific effects pose a challenge by potentially obscuring the biological signal and introducing unwanted variance. Existing harmonization techniques, which use statistical models to remove such effects, have been shown to incompletely remove site effects while also failing to preserve biological variability. More recently, generative models using GANs or autoencoder-based approaches, have been proposed for site adjustment. However, such methods are known for instability during training or blurry image generation. In recent years, diffusion models have become increasingly popular for their ability to generate high-quality synthetic images. In this work, we introduce the disentangled diffusion autoencoder (DDAE), a novel diffusion model designed for controlling specific aspects of an image. We apply the DDAE to the task of harmonizing MR images by generating high-quality site-adjusted images that preserve biological variability. We use data from 7 different sites and demonstrate the DDAE's superiority in generating high-resolution, harmonized 2D MR images over previous approaches. As far as we are aware, this work marks the first diffusion-based model for site adjustment of neuroimaging data.
Normative Diffusion Autoencoders: Application to Amyotrophic Lateral Sclerosis
Jul 19, 2024Abstract:Predicting survival in Amyotrophic Lateral Sclerosis (ALS) is a challenging task. Magnetic resonance imaging (MRI) data provide in vivo insight into brain health, but the low prevalence of the condition and resultant data scarcity limit training set sizes for prediction models. Survival models are further hindered by the subtle and often highly localised profile of ALS-related neurodegeneration. Normative models present a solution as they increase statistical power by leveraging large healthy cohorts. Separately, diffusion models excel in capturing the semantics embedded within images including subtle signs of accelerated brain ageing, which may help predict survival in ALS. Here, we combine the benefits of generative and normative modelling by introducing the normative diffusion autoencoder framework. To our knowledge, this is the first use of normative modelling within a diffusion autoencoder, as well as the first application of normative modelling to ALS. Our approach outperforms generative and non-generative normative modelling benchmarks in ALS prognostication, demonstrating enhanced predictive accuracy in the context of ALS survival prediction and normative modelling in general.
Semi-Supervised Diffusion Model for Brain Age Prediction
Feb 14, 2024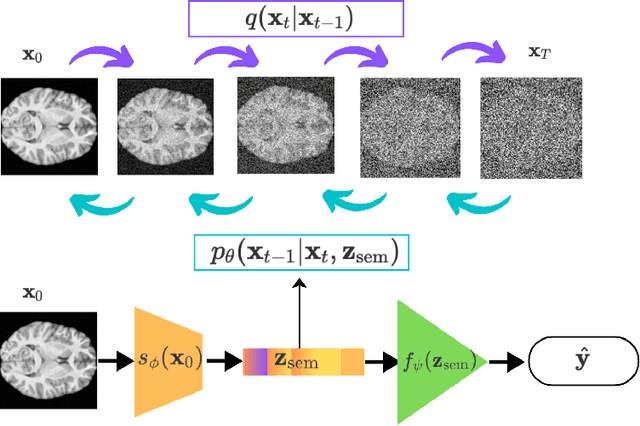

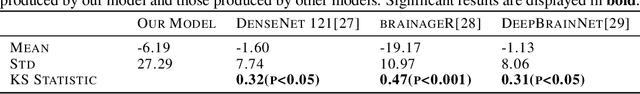
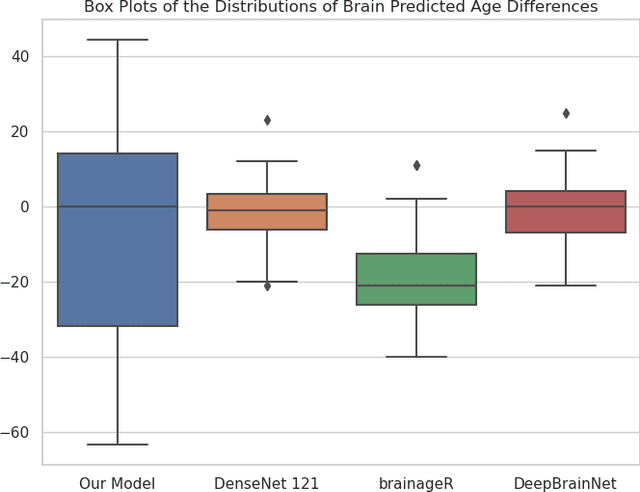
Abstract:Brain age prediction models have succeeded in predicting clinical outcomes in neurodegenerative diseases, but can struggle with tasks involving faster progressing diseases and low quality data. To enhance their performance, we employ a semi-supervised diffusion model, obtaining a 0.83(p<0.01) correlation between chronological and predicted age on low quality T1w MR images. This was competitive with state-of-the-art non-generative methods. Furthermore, the predictions produced by our model were significantly associated with survival length (r=0.24, p<0.05) in Amyotrophic Lateral Sclerosis. Thus, our approach demonstrates the value of diffusion-based architectures for the task of brain age prediction.
Interpretable Alzheimer's Disease Classification Via a Contrastive Diffusion Autoencoder
Jun 05, 2023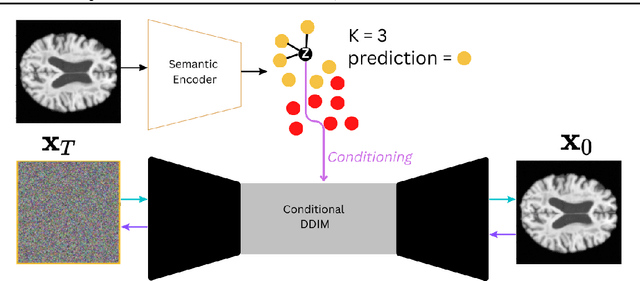
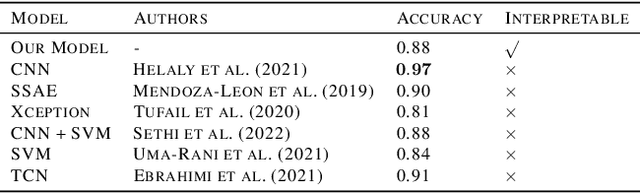
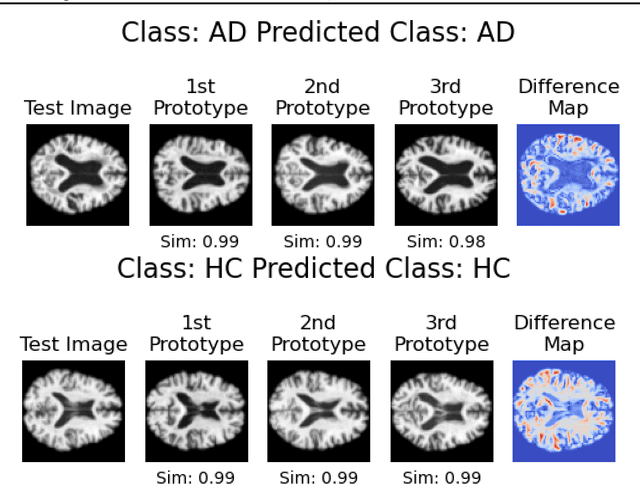
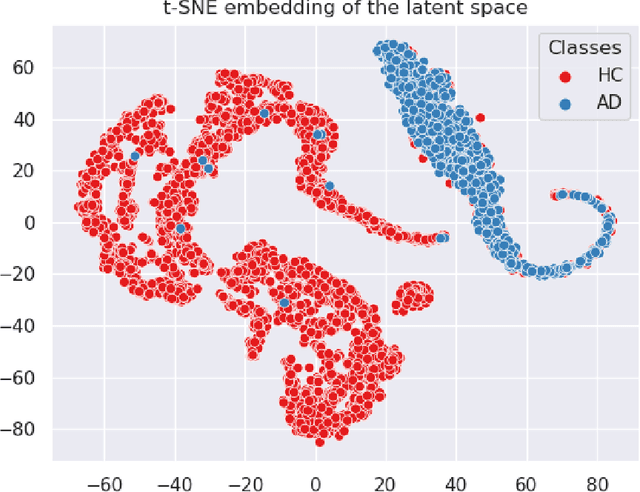
Abstract:In visual object classification, humans often justify their choices by comparing objects to prototypical examples within that class. We may therefore increase the interpretability of deep learning models by imbuing them with a similar style of reasoning. In this work, we apply this principle by classifying Alzheimer's Disease based on the similarity of images to training examples within the latent space. We use a contrastive loss combined with a diffusion autoencoder backbone, to produce a semantically meaningful latent space, such that neighbouring latents have similar image-level features. We achieve a classification accuracy comparable to black box approaches on a dataset of 2D MRI images, whilst producing human interpretable model explanations. Therefore, this work stands as a contribution to the pertinent development of accurate and interpretable deep learning within medical imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge