Stephen J. Price
Cross-modal Diffusion Modelling for Super-resolved Spatial Transcriptomics
Apr 19, 2024Abstract:The recent advancement of spatial transcriptomics (ST) allows to characterize spatial gene expression within tissue for discovery research. However, current ST platforms suffer from low resolution, hindering in-depth understanding of spatial gene expression. Super-resolution approaches promise to enhance ST maps by integrating histology images with gene expressions of profiled tissue spots. However, current super-resolution methods are limited by restoration uncertainty and mode collapse. Although diffusion models have shown promise in capturing complex interactions between multi-modal conditions, it remains a challenge to integrate histology images and gene expression for super-resolved ST maps. This paper proposes a cross-modal conditional diffusion model for super-resolving ST maps with the guidance of histology images. Specifically, we design a multi-modal disentangling network with cross-modal adaptive modulation to utilize complementary information from histology images and spatial gene expression. Moreover, we propose a dynamic cross-attention modelling strategy to extract hierarchical cell-to-tissue information from histology images. Lastly, we propose a co-expression-based gene-correlation graph network to model the co-expression relationship of multiple genes. Experiments show that our method outperforms other state-of-the-art methods in ST super-resolution on three public datasets.
Multi-modal learning for predicting the genotype of glioma
Mar 21, 2022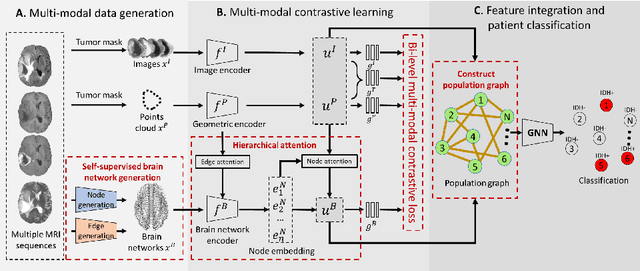
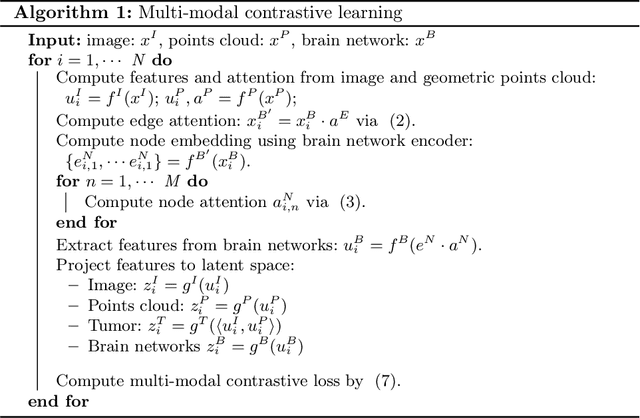
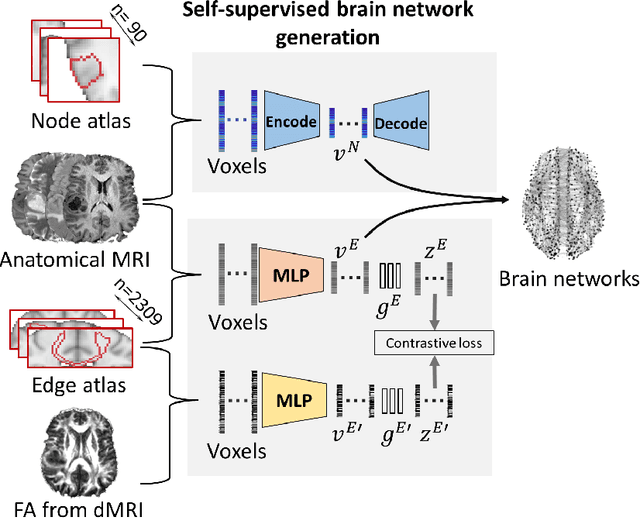

Abstract:The isocitrate dehydrogenase (IDH) gene mutation is an essential biomarker for the diagnosis and prognosis of glioma. It is promising to better predict glioma genotype by integrating focal tumor image and geometric features with brain network features derived from MRI. Convolutions neural networks show reasonable performance in predicting IDH mutation, which, however, cannot learn from non-Euclidean data, e.g., geometric and network data. In this study, we propose a multi-modal learning framework using three separate encoders to extract features of focal tumor image, tumor geometrics and global brain networks. To mitigate the limited availability of diffusion MRI, we develop a self-supervised approach to generate brain networks from anatomical multi-sequence MRI. Moreover, to extract tumor-related features from the brain network, we design a hierarchical attention module for the brain network encoder. Further, we design a bi-level multi-modal contrastive loss to align the multi-modal features and tackle the domain gap at the focal tumor and global brain. Finally, we propose a weighted population graph to integrate the multi-modal features for genotype prediction. Experimental results on the testing set show that the proposed model outperforms the baseline deep learning models. The ablation experiments validate the performance of different components of the framework. The visualized interpretation corresponds to clinical knowledge with further validation. In conclusion, the proposed learning framework provides a novel approach for predicting the genotype of glioma.
Predicting conversion of mild cognitive impairment to Alzheimer's disease
Mar 08, 2022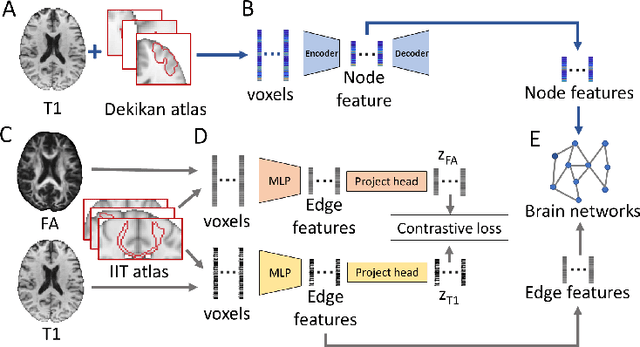
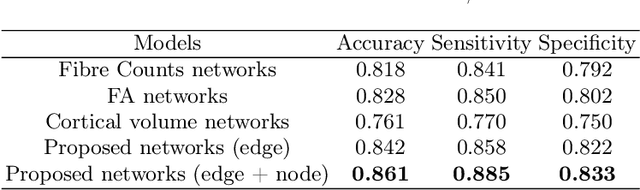
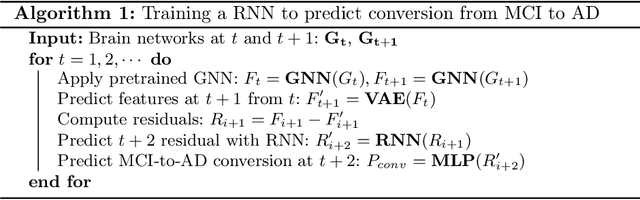
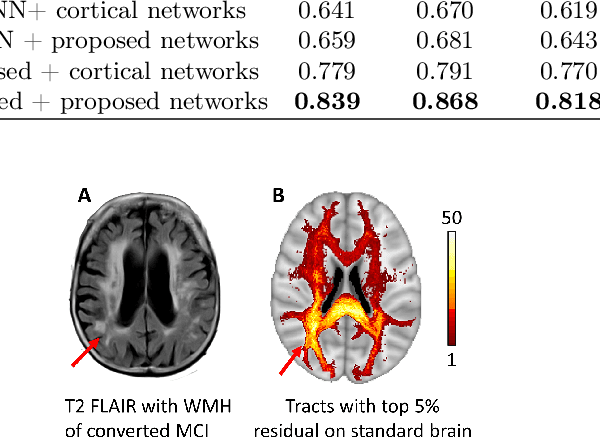
Abstract:Alzheimer's disease (AD) is the most common age-related dementia. Mild cognitive impairment (MCI) is the early stage of cognitive decline before AD. It is crucial to predict the MCI-to-AD conversion for precise management, which remains challenging due to the diversity of patients. Previous evidence shows that the brain network generated from diffusion MRI promises to classify dementia using deep learning. However, the limited availability of diffusion MRI challenges the model training. In this study, we develop a self-supervised contrastive learning approach to generate structural brain networks from routine anatomical MRI under the guidance of diffusion MRI. The generated brain networks are applied to train a learning framework for predicting the MCI-to-AD conversion. Instead of directly modelling the AD brain networks, we train a graph encoder and a variational autoencoder to model the healthy ageing trajectories from brain networks of healthy controls. To predict the MCI-to-AD conversion, we further design a recurrent neural networks based approach to model the longitudinal deviation of patients' brain networks from the healthy ageing trajectory. Numerical results show that the proposed methods outperform the benchmarks in the prediction task. We also visualize the model interpretation to explain the prediction and identify abnormal changes of white matter tracts.
Collaborative learning of images and geometrics for predicting isocitrate dehydrogenase status of glioma
Jan 14, 2022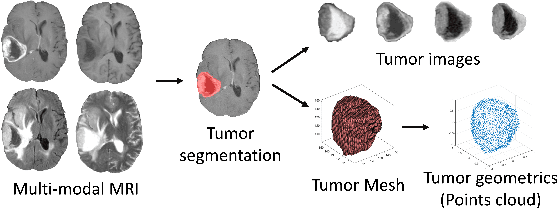

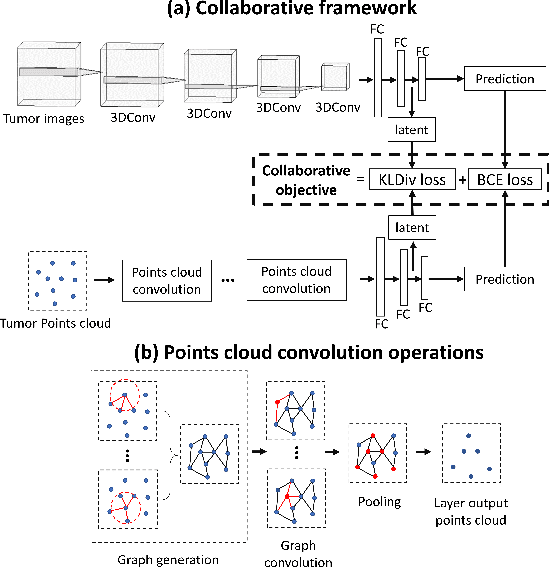
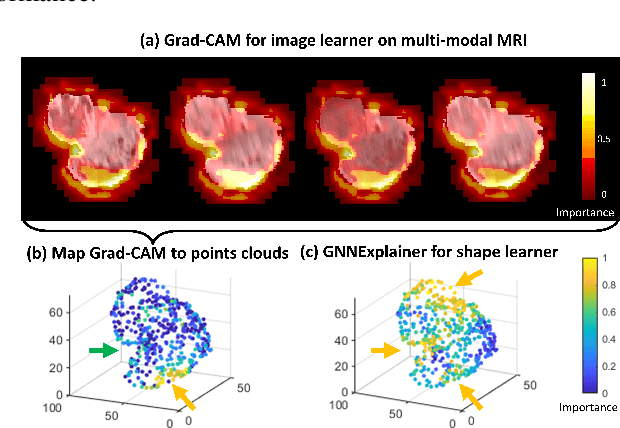
Abstract:The isocitrate dehydrogenase (IDH) gene mutation status is an important biomarker for glioma patients. The gold standard of IDH mutation detection requires tumour tissue obtained via invasive approaches and is usually expensive. Recent advancement in radiogenomics provides a non-invasive approach for predicting IDH mutation based on MRI. Meanwhile, tumor geometrics encompass crucial information for tumour phenotyping. Here we propose a collaborative learning framework that learns both tumor images and tumor geometrics using convolutional neural networks (CNN) and graph neural networks (GNN), respectively. Our results show that the proposed model outperforms the baseline model of 3D-DenseNet121. Further, the collaborative learning model achieves better performance than either the CNN or the GNN alone. The model interpretation shows that the CNN and GNN could identify common and unique regions of interest for IDH mutation prediction. In conclusion, collaborating image and geometric learners provides a novel approach for predicting genotype and characterising glioma.
Predicting isocitrate dehydrogenase mutation status in glioma using structural brain networks and graph neural networks
Sep 10, 2021Abstract:Glioma is a common malignant brain tumor with distinct survival among patients. The isocitrate dehydrogenase (IDH) gene mutation provides critical diagnostic and prognostic value for glioma. It is of crucial significance to non-invasively predict IDH mutation based on pre-treatment MRI. Machine learning/deep learning models show reasonable performance in predicting IDH mutation using MRI. However, most models neglect the systematic brain alterations caused by tumor invasion, where widespread infiltration along white matter tracts is a hallmark of glioma. Structural brain network provides an effective tool to characterize brain organisation, which could be captured by the graph neural networks (GNN) to more accurately predict IDH mutation. Here we propose a method to predict IDH mutation using GNN, based on the structural brain network of patients. Specifically, we firstly construct a network template of healthy subjects, consisting of atlases of edges (white matter tracts) and nodes (cortical/subcortical brain regions) to provide regions of interest (ROIs). Next, we employ autoencoders to extract the latent multi-modal MRI features from the ROIs of edges and nodes in patients, to train a GNN architecture for predicting IDH mutation. The results show that the proposed method outperforms the baseline models using the 3D-CNN and 3D-DenseNet. In addition, model interpretation suggests its ability to identify the tracts infiltrated by tumor, corresponding to clinical prior knowledge. In conclusion, integrating brain networks with GNN offers a new avenue to study brain lesions using computational neuroscience and computer vision approaches.
Expectation-Maximization Regularized Deep Learning for Weakly Supervised Tumor Segmentation for Glioblastoma
Jan 22, 2021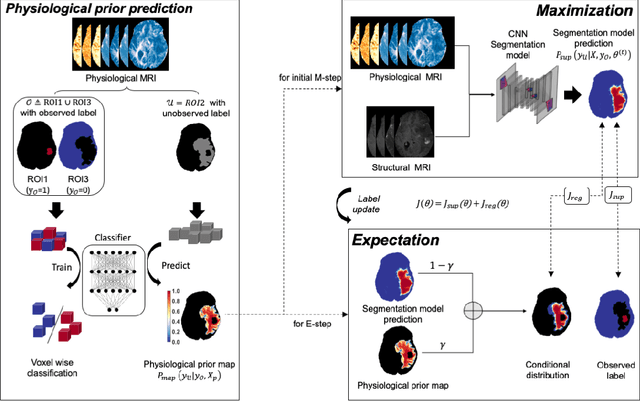
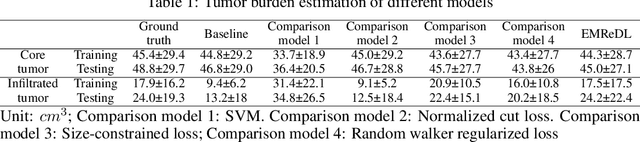
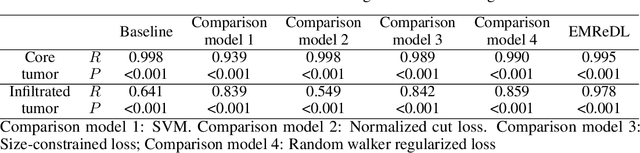
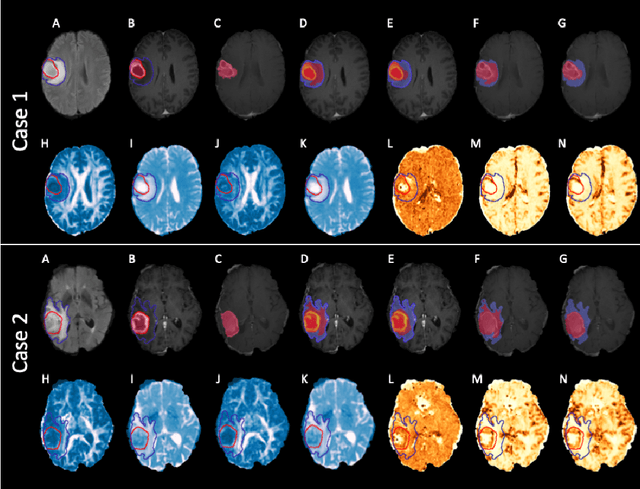
Abstract:We present an Expectation-Maximization (EM) Regularized Deep Learning (EMReDL) model for the weakly supervised tumor segmentation. The proposed framework was tailored to glioblastoma, a type of malignant tumor characterized by its diffuse infiltration into the surrounding brain tissue, which poses significant challenge to treatment target and tumor burden estimation based on conventional structural MRI. Although physiological MRI can provide more specific information regarding tumor infiltration, the relatively low resolution hinders a precise full annotation. This has motivated us to develop a weakly supervised deep learning solution that exploits the partial labelled tumor regions. EMReDL contains two components: a physiological prior prediction model and EM-regularized segmentation model. The physiological prior prediction model exploits the physiological MRI by training a classifier to generate a physiological prior map. This map was passed to the segmentation model for regularization using the EM algorithm. We evaluated the model on a glioblastoma dataset with the available pre-operative multiparametric MRI and recurrence MRI. EMReDL was shown to effectively segment the infiltrated tumor from the partially labelled region of potential infiltration. The segmented core and infiltrated tumor showed high consistency with the tumor burden labelled by experts. The performance comparison showed that EMReDL achieved higher accuracy than published state-of-the-art models. On MR spectroscopy, the segmented region showed more aggressive features than other partial labelled region. The proposed model can be generalized to other segmentation tasks with partial labels, with the CNN architecture flexible in the framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge