Guanwen Qiu
Reconstructing Cell Lineage Trees from Phenotypic Features with Metric Learning
Mar 18, 2025Abstract:How a single fertilized cell gives rise to a complex array of specialized cell types in development is a central question in biology. The cells grow, divide, and acquire differentiated characteristics through poorly understood molecular processes. A key approach to studying developmental processes is to infer the tree graph of cell lineage division and differentiation histories, providing an analytical framework for dissecting individual cells' molecular decisions during replication and differentiation. Although genetically engineered lineage-tracing methods have advanced the field, they are either infeasible or ethically constrained in many organisms. In contrast, modern single-cell technologies can measure high-content molecular profiles (e.g., transcriptomes) in a wide range of biological systems. Here, we introduce CellTreeQM, a novel deep learning method based on transformer architectures that learns an embedding space with geometric properties optimized for tree-graph inference. By formulating lineage reconstruction as a tree-metric learning problem, we have systematically explored supervised, weakly supervised, and unsupervised training settings and present a Lineage Reconstruction Benchmark to facilitate comprehensive evaluation of our learning method. We benchmarked the method on (1) synthetic data modeled via Brownian motion with independent noise and spurious signals and (2) lineage-resolved single-cell RNA sequencing datasets. Experimental results show that CellTreeQM recovers lineage structures with minimal supervision and limited data, offering a scalable framework for uncovering cell lineage relationships in challenging animal models. To our knowledge, this is the first method to cast cell lineage inference explicitly as a metric learning task, paving the way for future computational models aimed at uncovering the molecular dynamics of cell lineage.
Metastatic Cancer Image Classification Based On Deep Learning Method
Nov 13, 2020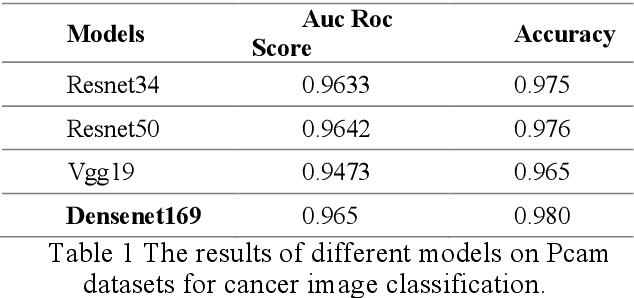
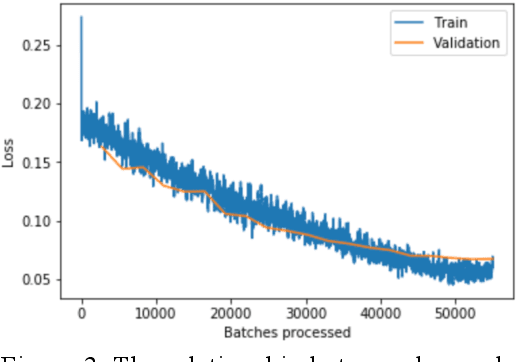
Abstract:Using histopathological images to automatically classify cancer is a difficult task for accurately detecting cancer, especially to identify metastatic cancer in small image patches obtained from larger digital pathology scans. Computer diagnosis technology has attracted wide attention from researchers. In this paper, we propose a noval method which combines the deep learning algorithm in image classification, the DenseNet169 framework and Rectified Adam optimization algorithm. The connectivity pattern of DenseNet is direct connections from any layer to all consecutive layers, which can effectively improve the information flow between different layers. With the fact that RAdam is not easy to fall into a local optimal solution, and it can converge quickly in model training. The experimental results shows that our model achieves superior performance over the other classical convolutional neural networks approaches, such as Vgg19, Resnet34, Resnet50. In particular, the Auc-Roc score of our DenseNet169 model is 1.77% higher than Vgg19 model, and the Accuracy score is 1.50% higher. Moreover, we also study the relationship between loss value and batches processed during the training stage and validation stage, and obtain some important and interesting findings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge