Boyang Zhao
DDNet: A Dual-Stream Graph Learning and Disentanglement Framework for Temporal Forgery Localization
Jan 05, 2026Abstract:The rapid evolution of AIGC technology enables misleading viewers by tampering mere small segments within a video, rendering video-level detection inaccurate and unpersuasive. Consequently, temporal forgery localization (TFL), which aims to precisely pinpoint tampered segments, becomes critical. However, existing methods are often constrained by \emph{local view}, failing to capture global anomalies. To address this, we propose a \underline{d}ual-stream graph learning and \underline{d}isentanglement framework for temporal forgery localization (DDNet). By coordinating a \emph{Temporal Distance Stream} for local artifacts and a \emph{Semantic Content Stream} for long-range connections, DDNet prevents global cues from being drowned out by local smoothness. Furthermore, we introduce Trace Disentanglement and Adaptation (TDA) to isolate generic forgery fingerprints, alongside Cross-Level Feature Embedding (CLFE) to construct a robust feature foundation via deep fusion of hierarchical features. Experiments on ForgeryNet and TVIL benchmarks demonstrate that our method outperforms state-of-the-art approaches by approximately 9\% in AP@0.95, with significant improvements in cross-domain robustness.
LiDARDustX: A LiDAR Dataset for Dusty Unstructured Road Environments
May 28, 2025



Abstract:Autonomous driving datasets are essential for validating the progress of intelligent vehicle algorithms, which include localization, perception, and prediction. However, existing datasets are predominantly focused on structured urban environments, which limits the exploration of unstructured and specialized scenarios, particularly those characterized by significant dust levels. This paper introduces the LiDARDustX dataset, which is specifically designed for perception tasks under high-dust conditions, such as those encountered in mining areas. The LiDARDustX dataset consists of 30,000 LiDAR frames captured by six different LiDAR sensors, each accompanied by 3D bounding box annotations and point cloud semantic segmentation. Notably, over 80% of the dataset comprises dust-affected scenes. By utilizing this dataset, we have established a benchmark for evaluating the performance of state-of-the-art 3D detection and segmentation algorithms. Additionally, we have analyzed the impact of dust on perception accuracy and delved into the causes of these effects. The data and further information can be accessed at: https://github.com/vincentweikey/LiDARDustX.
Joint Modelling Histology and Molecular Markers for Cancer Classification
Feb 11, 2025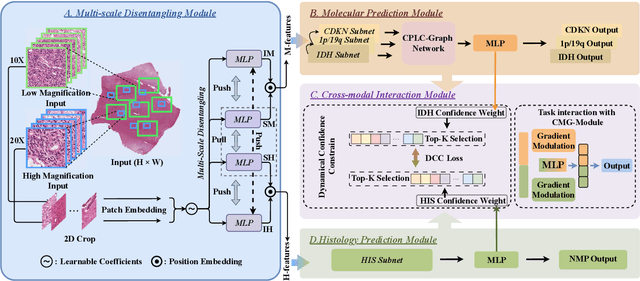
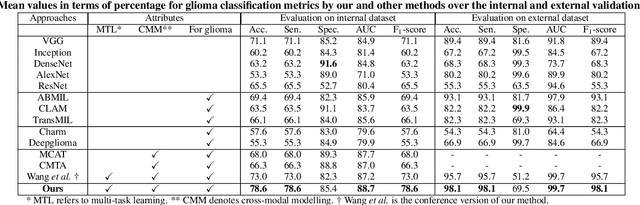
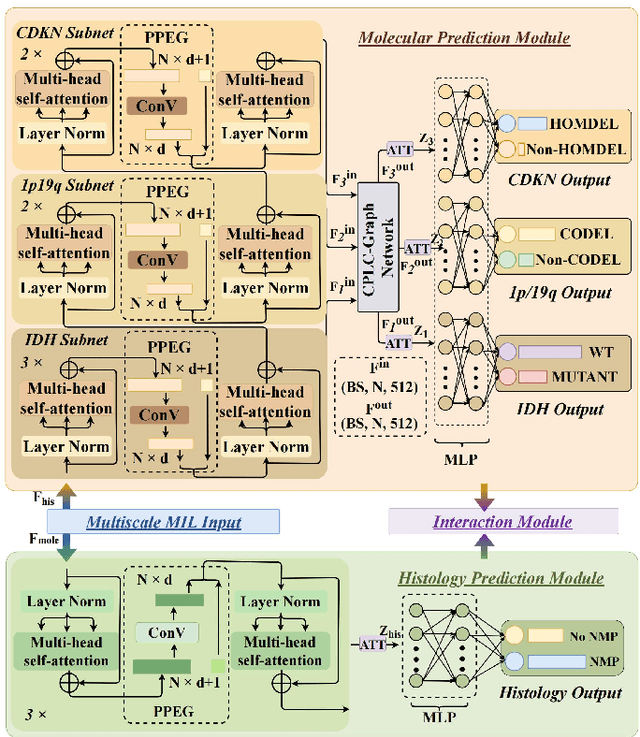
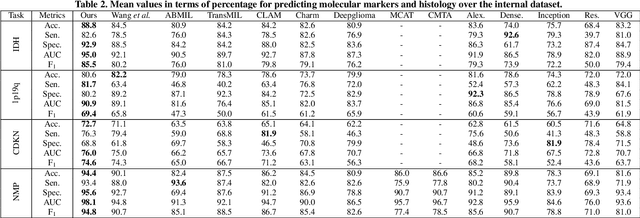
Abstract:Cancers are characterized by remarkable heterogeneity and diverse prognosis. Accurate cancer classification is essential for patient stratification and clinical decision-making. Although digital pathology has been advancing cancer diagnosis and prognosis, the paradigm in cancer pathology has shifted from purely relying on histology features to incorporating molecular markers. There is an urgent need for digital pathology methods to meet the needs of the new paradigm. We introduce a novel digital pathology approach to jointly predict molecular markers and histology features and model their interactions for cancer classification. Firstly, to mitigate the challenge of cross-magnification information propagation, we propose a multi-scale disentangling module, enabling the extraction of multi-scale features from high-magnification (cellular-level) to low-magnification (tissue-level) whole slide images. Further, based on the multi-scale features, we propose an attention-based hierarchical multi-task multi-instance learning framework to simultaneously predict histology and molecular markers. Moreover, we propose a co-occurrence probability-based label correlation graph network to model the co-occurrence of molecular markers. Lastly, we design a cross-modal interaction module with the dynamic confidence constrain loss and a cross-modal gradient modulation strategy, to model the interactions of histology and molecular markers. Our experiments demonstrate that our method outperforms other state-of-the-art methods in classifying glioma, histology features and molecular markers. Our method promises to promote precise oncology with the potential to advance biomedical research and clinical applications. The code is available at https://github.com/LHY1007/M3C2
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge