Fangliangzi Meng
Knowledge-driven Subspace Fusion and Gradient Coordination for Multi-modal Learning
Jun 20, 2024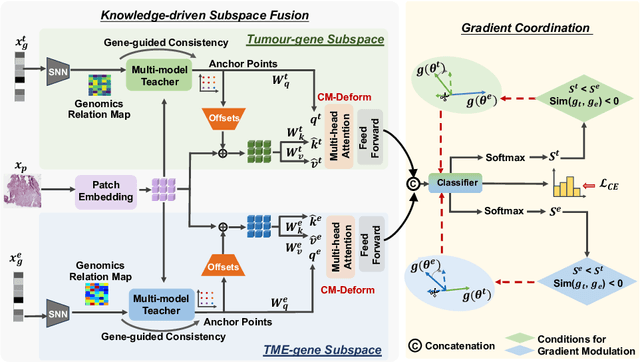
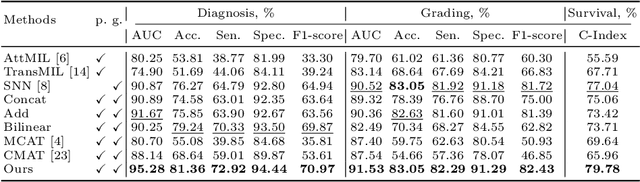

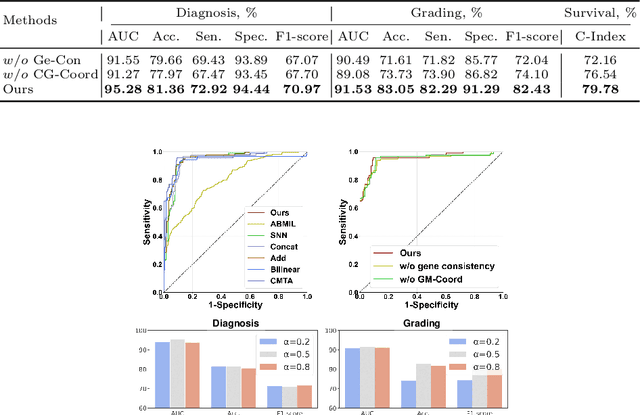
Abstract:Multi-modal learning plays a crucial role in cancer diagnosis and prognosis. Current deep learning based multi-modal approaches are often limited by their abilities to model the complex correlations between genomics and histology data, addressing the intrinsic complexity of tumour ecosystem where both tumour and microenvironment contribute to malignancy. We propose a biologically interpretative and robust multi-modal learning framework to efficiently integrate histology images and genomics by decomposing the feature subspace of histology images and genomics, reflecting distinct tumour and microenvironment features. To enhance cross-modal interactions, we design a knowledge-driven subspace fusion scheme, consisting of a cross-modal deformable attention module and a gene-guided consistency strategy. Additionally, in pursuit of dynamically optimizing the subspace knowledge, we further propose a novel gradient coordination learning strategy. Extensive experiments demonstrate the effectiveness of the proposed method, outperforming state-of-the-art techniques in three downstream tasks of glioma diagnosis, tumour grading, and survival analysis. Our code is available at https://github.com/helenypzhang/Subspace-Multimodal-Learning.
Genomics-guided Representation Learning for Pathologic Pan-cancer Tumor Microenvironment Subtype Prediction
Jun 10, 2024



Abstract:The characterization of Tumor MicroEnvironment (TME) is challenging due to its complexity and heterogeneity. Relatively consistent TME characteristics embedded within highly specific tissue features, render them difficult to predict. The capability to accurately classify TME subtypes is of critical significance for clinical tumor diagnosis and precision medicine. Based on the observation that tumors with different origins share similar microenvironment patterns, we propose PathoTME, a genomics-guided Siamese representation learning framework employing Whole Slide Image (WSI) for pan-cancer TME subtypes prediction. Specifically, we utilize Siamese network to leverage genomic information as a regularization factor to assist WSI embeddings learning during the training phase. Additionally, we employ Domain Adversarial Neural Network (DANN) to mitigate the impact of tissue type variations. To eliminate domain bias, a dynamic WSI prompt is designed to further unleash the model's capabilities. Our model achieves better performance than other state-of-the-art methods across 23 cancer types on TCGA dataset. Our code is available at https://github.com/Mengflz/PathoTME.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge