Xianjin Dai
Large Language Model-Augmented Auto-Delineation of Treatment Target Volume in Radiation Therapy
Jul 10, 2024Abstract:Radiation therapy (RT) is one of the most effective treatments for cancer, and its success relies on the accurate delineation of targets. However, target delineation is a comprehensive medical decision that currently relies purely on manual processes by human experts. Manual delineation is time-consuming, laborious, and subject to interobserver variations. Although the advancements in artificial intelligence (AI) techniques have significantly enhanced the auto-contouring of normal tissues, accurate delineation of RT target volumes remains a challenge. In this study, we propose a visual language model-based RT target volume auto-delineation network termed Radformer. The Radformer utilizes a hierarichal vision transformer as the backbone and incorporates large language models to extract text-rich features from clinical data. We introduce a visual language attention module (VLAM) for integrating visual and linguistic features for language-aware visual encoding (LAVE). The Radformer has been evaluated on a dataset comprising 2985 patients with head-and-neck cancer who underwent RT. Metrics, including the Dice similarity coefficient (DSC), intersection over union (IOU), and 95th percentile Hausdorff distance (HD95), were used to evaluate the performance of the model quantitatively. Our results demonstrate that the Radformer has superior segmentation performance compared to other state-of-the-art models, validating its potential for adoption in RT practice.
Landmark Tracking in Liver US images Using Cascade Convolutional Neural Networks with Long Short-Term Memory
Sep 14, 2022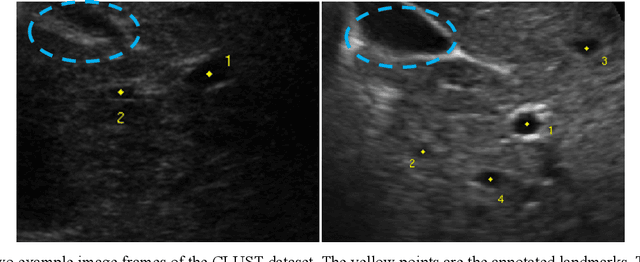
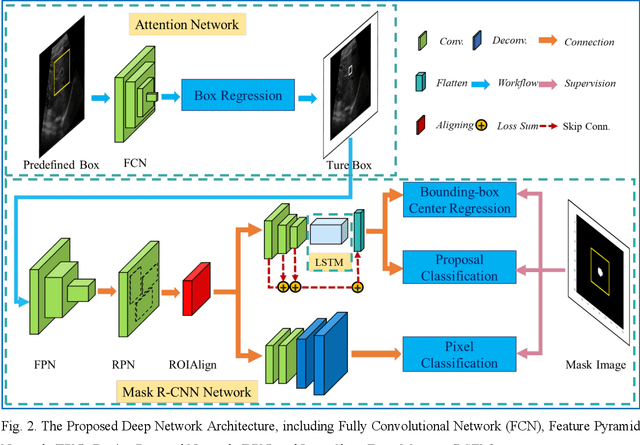
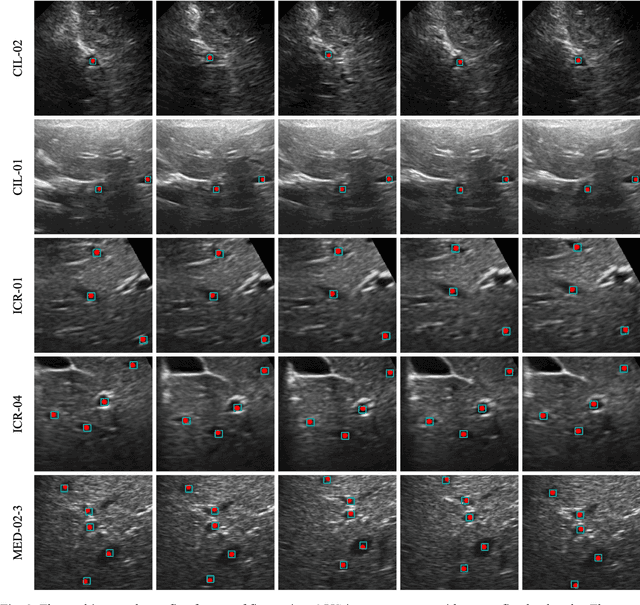
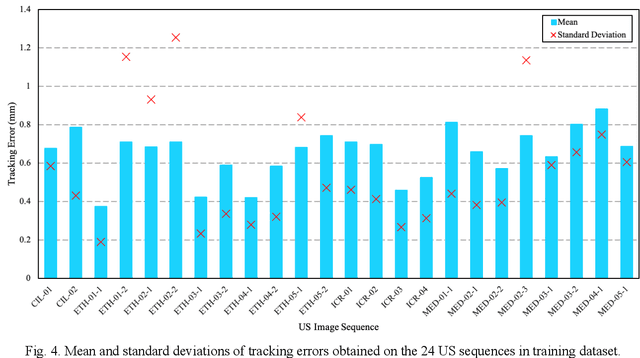
Abstract:This study proposed a deep learning-based tracking method for ultrasound (US) image-guided radiation therapy. The proposed cascade deep learning model is composed of an attention network, a mask region-based convolutional neural network (mask R-CNN), and a long short-term memory (LSTM) network. The attention network learns a mapping from a US image to a suspected area of landmark motion in order to reduce the search region. The mask R-CNN then produces multiple region-of-interest (ROI) proposals in the reduced region and identifies the proposed landmark via three network heads: bounding box regression, proposal classification, and landmark segmentation. The LSTM network models the temporal relationship among the successive image frames for bounding box regression and proposal classification. To consolidate the final proposal, a selection method is designed according to the similarities between sequential frames. The proposed method was tested on the liver US tracking datasets used in the Medical Image Computing and Computer Assisted Interventions (MICCAI) 2015 challenges, where the landmarks were annotated by three experienced observers to obtain their mean positions. Five-fold cross-validation on the 24 given US sequences with ground truths shows that the mean tracking error for all landmarks is 0.65+/-0.56 mm, and the errors of all landmarks are within 2 mm. We further tested the proposed model on 69 landmarks from the testing dataset that has a similar image pattern to the training pattern, resulting in a mean tracking error of 0.94+/-0.83 mm. Our experimental results have demonstrated the feasibility and accuracy of our proposed method in tracking liver anatomic landmarks using US images, providing a potential solution for real-time liver tracking for active motion management during radiation therapy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge