Lingcong Cai
VisionLLM-based Multimodal Fusion Network for Glottic Carcinoma Early Detection
Dec 24, 2024Abstract:The early detection of glottic carcinoma is critical for improving patient outcomes, as it enables timely intervention, preserves vocal function, and significantly reduces the risk of tumor progression and metastasis. However, the similarity in morphology between glottic carcinoma and vocal cord dysplasia results in suboptimal detection accuracy. To address this issue, we propose a vision large language model-based (VisionLLM-based) multimodal fusion network for glottic carcinoma detection, known as MMGC-Net. By integrating image and text modalities, multimodal models can capture complementary information, leading to more accurate and robust predictions. In this paper, we collect a private real glottic carcinoma dataset named SYSU1H from the First Affiliated Hospital of Sun Yat-sen University, with 5,799 image-text pairs. We leverage an image encoder and additional Q-Former to extract vision embeddings and the Large Language Model Meta AI (Llama3) to obtain text embeddings. These modalities are then integrated through a laryngeal feature fusion block, enabling a comprehensive integration of image and text features, thereby improving the glottic carcinoma identification performance. Extensive experiments on the SYSU1H dataset demonstrate that MMGC-Net can achieve state-of-the-art performance, which is superior to previous multimodal models.
Stain-aware Domain Alignment for Imbalance Blood Cell Classification
Dec 04, 2024



Abstract:Blood cell identification is critical for hematological analysis as it aids physicians in diagnosing various blood-related diseases. In real-world scenarios, blood cell image datasets often present the issues of domain shift and data imbalance, posing challenges for accurate blood cell identification. To address these issues, we propose a novel blood cell classification method termed SADA via stain-aware domain alignment. The primary objective of this work is to mine domain-invariant features in the presence of domain shifts and data imbalances. To accomplish this objective, we propose a stain-based augmentation approach and a local alignment constraint to learn domain-invariant features. Furthermore, we propose a domain-invariant supervised contrastive learning strategy to capture discriminative features. We decouple the training process into two stages of domain-invariant feature learning and classification training, alleviating the problem of data imbalance. Experiment results on four public blood cell datasets and a private real dataset collected from the Third Affiliated Hospital of Sun Yat-sen University demonstrate that SADA can achieve a new state-of-the-art baseline, which is superior to the existing cutting-edge methods with a big margin. The source code can be available at the URL (\url{https://github.com/AnoK3111/SADA}).
LoCo: Low-Contrast-Enhanced Contrastive Learning for Semi-Supervised Endoscopic Image Segmentation
Dec 03, 2024Abstract:The segmentation of endoscopic images plays a vital role in computer-aided diagnosis and treatment. The advancements in deep learning have led to the employment of numerous models for endoscopic tumor segmentation, achieving promising segmentation performance. Despite recent advancements, precise segmentation remains challenging due to limited annotations and the issue of low contrast. To address these issues, we propose a novel semi-supervised segmentation framework termed LoCo via low-contrast-enhanced contrastive learning (LCC). This innovative approach effectively harnesses the vast amounts of unlabeled data available for endoscopic image segmentation, improving both accuracy and robustness in the segmentation process. Specifically, LCC incorporates two advanced strategies to enhance the distinctiveness of low-contrast pixels: inter-class contrast enhancement (ICE) and boundary contrast enhancement (BCE), enabling models to segment low-contrast pixels among malignant tumors, benign tumors, and normal tissues. Additionally, a confidence-based dynamic filter (CDF) is designed for pseudo-label selection, enhancing the utilization of generated pseudo-labels for unlabeled data with a specific focus on minority classes. Extensive experiments conducted on two public datasets, as well as a large proprietary dataset collected over three years, demonstrate that LoCo achieves state-of-the-art results, significantly outperforming previous methods. The source code of LoCo is available at the URL of https://github.com/AnoK3111/LoCo.
Domain-invariant Representation Learning via Segment Anything Model for Blood Cell Classification
Aug 14, 2024Abstract:Accurate classification of blood cells is of vital significance in the diagnosis of hematological disorders. However, in real-world scenarios, domain shifts caused by the variability in laboratory procedures and settings, result in a rapid deterioration of the model's generalization performance. To address this issue, we propose a novel framework of domain-invariant representation learning (DoRL) via segment anything model (SAM) for blood cell classification. The DoRL comprises two main components: a LoRA-based SAM (LoRA-SAM) and a cross-domain autoencoder (CAE). The advantage of DoRL is that it can extract domain-invariant representations from various blood cell datasets in an unsupervised manner. Specifically, we first leverage the large-scale foundation model of SAM, fine-tuned with LoRA, to learn general image embeddings and segment blood cells. Additionally, we introduce CAE to learn domain-invariant representations across different-domain datasets while mitigating images' artifacts. To validate the effectiveness of domain-invariant representations, we employ five widely used machine learning classifiers to construct blood cell classification models. Experimental results on two public blood cell datasets and a private real dataset demonstrate that our proposed DoRL achieves a new state-of-the-art cross-domain performance, surpassing existing methods by a significant margin. The source code can be available at the URL (https://github.com/AnoK3111/DoRL).
Towards Cross-Domain Single Blood Cell Image Classification via Large-Scale LoRA-based Segment Anything Model
Aug 13, 2024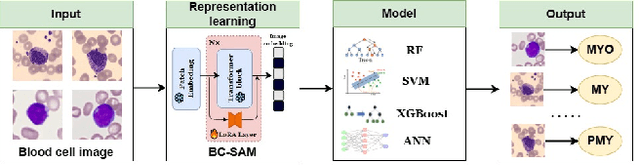
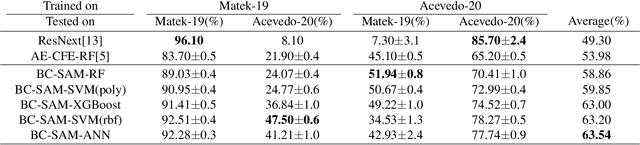
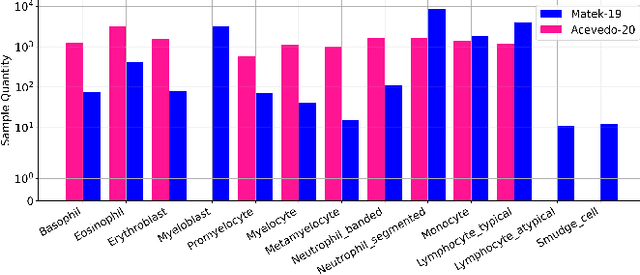
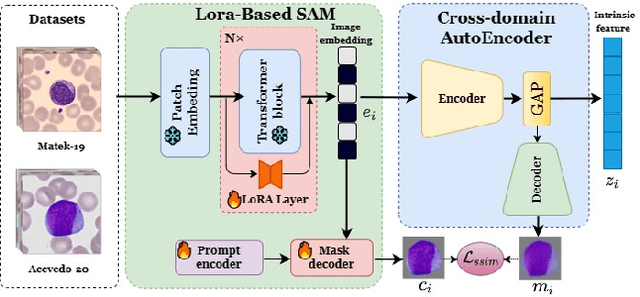
Abstract:Accurate classification of blood cells plays a vital role in hematological analysis as it aids physicians in diagnosing various medical conditions. In this study, we present a novel approach for classifying blood cell images known as BC-SAM. BC-SAM leverages the large-scale foundation model of Segment Anything Model (SAM) and incorporates a fine-tuning technique using LoRA, allowing it to extract general image embeddings from blood cell images. To enhance the applicability of BC-SAM across different blood cell image datasets, we introduce an unsupervised cross-domain autoencoder that focuses on learning intrinsic features while suppressing artifacts in the images. To assess the performance of BC-SAM, we employ four widely used machine learning classifiers (Random Forest, Support Vector Machine, Artificial Neural Network, and XGBoost) to construct blood cell classification models and compare them against existing state-of-the-art methods. Experimental results conducted on two publicly available blood cell datasets (Matek-19 and Acevedo-20) demonstrate that our proposed BC-SAM achieves a new state-of-the-art result, surpassing the baseline methods with a significant improvement. The source code of this paper is available at https://github.com/AnoK3111/BC-SAM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge