Xiaomao Fan
Multimodal Medical Endoscopic Image Analysis via Progressive Disentangle-aware Contrastive Learning
Aug 23, 2025

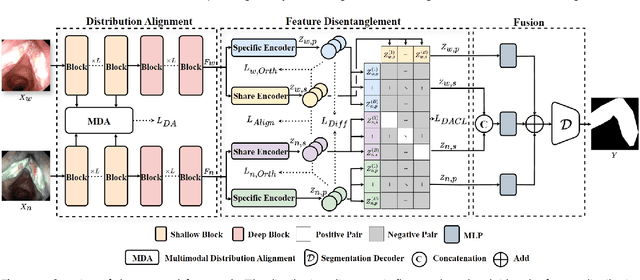

Abstract:Accurate segmentation of laryngo-pharyngeal tumors is crucial for precise diagnosis and effective treatment planning. However, traditional single-modality imaging methods often fall short of capturing the complex anatomical and pathological features of these tumors. In this study, we present an innovative multi-modality representation learning framework based on the `Align-Disentangle-Fusion' mechanism that seamlessly integrates 2D White Light Imaging (WLI) and Narrow Band Imaging (NBI) pairs to enhance segmentation performance. A cornerstone of our approach is multi-scale distribution alignment, which mitigates modality discrepancies by aligning features across multiple transformer layers. Furthermore, a progressive feature disentanglement strategy is developed with the designed preliminary disentanglement and disentangle-aware contrastive learning to effectively separate modality-specific and shared features, enabling robust multimodal contrastive learning and efficient semantic fusion. Comprehensive experiments on multiple datasets demonstrate that our method consistently outperforms state-of-the-art approaches, achieving superior accuracy across diverse real clinical scenarios.
ITCFN: Incomplete Triple-Modal Co-Attention Fusion Network for Mild Cognitive Impairment Conversion Prediction
Jan 20, 2025


Abstract:Alzheimer's disease (AD) is a common neurodegenerative disease among the elderly. Early prediction and timely intervention of its prodromal stage, mild cognitive impairment (MCI), can decrease the risk of advancing to AD. Combining information from various modalities can significantly improve predictive accuracy. However, challenges such as missing data and heterogeneity across modalities complicate multimodal learning methods as adding more modalities can worsen these issues. Current multimodal fusion techniques often fail to adapt to the complexity of medical data, hindering the ability to identify relationships between modalities. To address these challenges, we propose an innovative multimodal approach for predicting MCI conversion, focusing specifically on the issues of missing positron emission tomography (PET) data and integrating diverse medical information. The proposed incomplete triple-modal MCI conversion prediction network is tailored for this purpose. Through the missing modal generation module, we synthesize the missing PET data from the magnetic resonance imaging and extract features using specifically designed encoders. We also develop a channel aggregation module and a triple-modal co-attention fusion module to reduce feature redundancy and achieve effective multimodal data fusion. Furthermore, we design a loss function to handle missing modality issues and align cross-modal features. These components collectively harness multimodal data to boost network performance. Experimental results on the ADNI1 and ADNI2 datasets show that our method significantly surpasses existing unimodal and other multimodal models. Our code is available at https://github.com/justinhxy/ITFC.
VisionLLM-based Multimodal Fusion Network for Glottic Carcinoma Early Detection
Dec 24, 2024Abstract:The early detection of glottic carcinoma is critical for improving patient outcomes, as it enables timely intervention, preserves vocal function, and significantly reduces the risk of tumor progression and metastasis. However, the similarity in morphology between glottic carcinoma and vocal cord dysplasia results in suboptimal detection accuracy. To address this issue, we propose a vision large language model-based (VisionLLM-based) multimodal fusion network for glottic carcinoma detection, known as MMGC-Net. By integrating image and text modalities, multimodal models can capture complementary information, leading to more accurate and robust predictions. In this paper, we collect a private real glottic carcinoma dataset named SYSU1H from the First Affiliated Hospital of Sun Yat-sen University, with 5,799 image-text pairs. We leverage an image encoder and additional Q-Former to extract vision embeddings and the Large Language Model Meta AI (Llama3) to obtain text embeddings. These modalities are then integrated through a laryngeal feature fusion block, enabling a comprehensive integration of image and text features, thereby improving the glottic carcinoma identification performance. Extensive experiments on the SYSU1H dataset demonstrate that MMGC-Net can achieve state-of-the-art performance, which is superior to previous multimodal models.
Stain-aware Domain Alignment for Imbalance Blood Cell Classification
Dec 04, 2024



Abstract:Blood cell identification is critical for hematological analysis as it aids physicians in diagnosing various blood-related diseases. In real-world scenarios, blood cell image datasets often present the issues of domain shift and data imbalance, posing challenges for accurate blood cell identification. To address these issues, we propose a novel blood cell classification method termed SADA via stain-aware domain alignment. The primary objective of this work is to mine domain-invariant features in the presence of domain shifts and data imbalances. To accomplish this objective, we propose a stain-based augmentation approach and a local alignment constraint to learn domain-invariant features. Furthermore, we propose a domain-invariant supervised contrastive learning strategy to capture discriminative features. We decouple the training process into two stages of domain-invariant feature learning and classification training, alleviating the problem of data imbalance. Experiment results on four public blood cell datasets and a private real dataset collected from the Third Affiliated Hospital of Sun Yat-sen University demonstrate that SADA can achieve a new state-of-the-art baseline, which is superior to the existing cutting-edge methods with a big margin. The source code can be available at the URL (\url{https://github.com/AnoK3111/SADA}).
LoCo: Low-Contrast-Enhanced Contrastive Learning for Semi-Supervised Endoscopic Image Segmentation
Dec 03, 2024Abstract:The segmentation of endoscopic images plays a vital role in computer-aided diagnosis and treatment. The advancements in deep learning have led to the employment of numerous models for endoscopic tumor segmentation, achieving promising segmentation performance. Despite recent advancements, precise segmentation remains challenging due to limited annotations and the issue of low contrast. To address these issues, we propose a novel semi-supervised segmentation framework termed LoCo via low-contrast-enhanced contrastive learning (LCC). This innovative approach effectively harnesses the vast amounts of unlabeled data available for endoscopic image segmentation, improving both accuracy and robustness in the segmentation process. Specifically, LCC incorporates two advanced strategies to enhance the distinctiveness of low-contrast pixels: inter-class contrast enhancement (ICE) and boundary contrast enhancement (BCE), enabling models to segment low-contrast pixels among malignant tumors, benign tumors, and normal tissues. Additionally, a confidence-based dynamic filter (CDF) is designed for pseudo-label selection, enhancing the utilization of generated pseudo-labels for unlabeled data with a specific focus on minority classes. Extensive experiments conducted on two public datasets, as well as a large proprietary dataset collected over three years, demonstrate that LoCo achieves state-of-the-art results, significantly outperforming previous methods. The source code of LoCo is available at the URL of https://github.com/AnoK3111/LoCo.
SAM-Swin: SAM-Driven Dual-Swin Transformers with Adaptive Lesion Enhancement for Laryngo-Pharyngeal Tumor Detection
Oct 29, 2024Abstract:Laryngo-pharyngeal cancer (LPC) is a highly lethal malignancy in the head and neck region. Recent advancements in tumor detection, particularly through dual-branch network architectures, have significantly improved diagnostic accuracy by integrating global and local feature extraction. However, challenges remain in accurately localizing lesions and fully capitalizing on the complementary nature of features within these branches. To address these issues, we propose SAM-Swin, an innovative SAM-driven Dual-Swin Transformer for laryngo-pharyngeal tumor detection. This model leverages the robust segmentation capabilities of the Segment Anything Model 2 (SAM2) to achieve precise lesion segmentation. Meanwhile, we present a multi-scale lesion-aware enhancement module (MS-LAEM) designed to adaptively enhance the learning of nuanced complementary features across various scales, improving the quality of feature extraction and representation. Furthermore, we implement a multi-scale class-aware guidance (CAG) loss that delivers multi-scale targeted supervision, thereby enhancing the model's capacity to extract class-specific features. To validate our approach, we compiled three LPC datasets from the First Affiliated Hospital (FAHSYSU), the Sixth Affiliated Hospital (SAHSYSU) of Sun Yat-sen University, and Nanfang Hospital of Southern Medical University (NHSMU). The FAHSYSU dataset is utilized for internal training, while the SAHSYSU and NHSMU datasets serve for external evaluation. Extensive experiments demonstrate that SAM-Swin outperforms state-of-the-art methods, showcasing its potential for advancing LPC detection and improving patient outcomes. The source code of SAM-Swin is available at the URL of \href{https://github.com/VVJia/SAM-Swin}{https://github.com/VVJia/SAM-Swin}.
Mamba-Enhanced Text-Audio-Video Alignment Network for Emotion Recognition in Conversations
Sep 08, 2024



Abstract:Emotion Recognition in Conversations (ERCs) is a vital area within multimodal interaction research, dedicated to accurately identifying and classifying the emotions expressed by speakers throughout a conversation. Traditional ERC approaches predominantly rely on unimodal cues\-such as text, audio, or visual data\-leading to limitations in their effectiveness. These methods encounter two significant challenges: 1) Consistency in multimodal information. Before integrating various modalities, it is crucial to ensure that the data from different sources is aligned and coherent. 2) Contextual information capture. Successfully fusing multimodal features requires a keen understanding of the evolving emotional tone, especially in lengthy dialogues where emotions may shift and develop over time. To address these limitations, we propose a novel Mamba-enhanced Text-Audio-Video alignment network (MaTAV) for the ERC task. MaTAV is with the advantages of aligning unimodal features to ensure consistency across different modalities and handling long input sequences to better capture contextual multimodal information. The extensive experiments on the MELD and IEMOCAP datasets demonstrate that MaTAV significantly outperforms existing state-of-the-art methods on the ERC task with a big margin.
3D-LSPTM: An Automatic Framework with 3D-Large-Scale Pretrained Model for Laryngeal Cancer Detection Using Laryngoscopic Videos
Sep 02, 2024Abstract:Laryngeal cancer is a malignant disease with a high morality rate in otorhinolaryngology, posing an significant threat to human health. Traditionally larygologists manually visual-inspect laryngeal cancer in laryngoscopic videos, which is quite time-consuming and subjective. In this study, we propose a novel automatic framework via 3D-large-scale pretrained models termed 3D-LSPTM for laryngeal cancer detection. Firstly, we collect 1,109 laryngoscopic videos from the First Affiliated Hospital Sun Yat-sen University with the approval of the Ethics Committee. Then we utilize the 3D-large-scale pretrained models of C3D, TimeSformer, and Video-Swin-Transformer, with the merit of advanced featuring videos, for laryngeal cancer detection with fine-tuning techniques. Extensive experiments show that our proposed 3D-LSPTM can achieve promising performance on the task of laryngeal cancer detection. Particularly, 3D-LSPTM with the backbone of Video-Swin-Transformer can achieve 92.4% accuracy, 95.6% sensitivity, 94.1% precision, and 94.8% F_1.
SAM-FNet: SAM-Guided Fusion Network for Laryngo-Pharyngeal Tumor Detection
Aug 15, 2024Abstract:Laryngo-pharyngeal cancer (LPC) is a highly fatal malignant disease affecting the head and neck region. Previous studies on endoscopic tumor detection, particularly those leveraging dual-branch network architectures, have shown significant advancements in tumor detection. These studies highlight the potential of dual-branch networks in improving diagnostic accuracy by effectively integrating global and local (lesion) feature extraction. However, they are still limited in their capabilities to accurately locate the lesion region and capture the discriminative feature information between the global and local branches. To address these issues, we propose a novel SAM-guided fusion network (SAM-FNet), a dual-branch network for laryngo-pharyngeal tumor detection. By leveraging the powerful object segmentation capabilities of the Segment Anything Model (SAM), we introduce the SAM into the SAM-FNet to accurately segment the lesion region. Furthermore, we propose a GAN-like feature optimization (GFO) module to capture the discriminative features between the global and local branches, enhancing the fusion feature complementarity. Additionally, we collect two LPC datasets from the First Affiliated Hospital (FAHSYSU) and the Sixth Affiliated Hospital (SAHSYSU) of Sun Yat-sen University. The FAHSYSU dataset is used as the internal dataset for training the model, while the SAHSYSU dataset is used as the external dataset for evaluating the model's performance. Extensive experiments on both datasets of FAHSYSU and SAHSYSU demonstrate that the SAM-FNet can achieve competitive results, outperforming the state-of-the-art counterparts. The source code of SAM-FNet is available at the URL of https://github.com/VVJia/SAM-FNet.
Domain-invariant Representation Learning via Segment Anything Model for Blood Cell Classification
Aug 14, 2024Abstract:Accurate classification of blood cells is of vital significance in the diagnosis of hematological disorders. However, in real-world scenarios, domain shifts caused by the variability in laboratory procedures and settings, result in a rapid deterioration of the model's generalization performance. To address this issue, we propose a novel framework of domain-invariant representation learning (DoRL) via segment anything model (SAM) for blood cell classification. The DoRL comprises two main components: a LoRA-based SAM (LoRA-SAM) and a cross-domain autoencoder (CAE). The advantage of DoRL is that it can extract domain-invariant representations from various blood cell datasets in an unsupervised manner. Specifically, we first leverage the large-scale foundation model of SAM, fine-tuned with LoRA, to learn general image embeddings and segment blood cells. Additionally, we introduce CAE to learn domain-invariant representations across different-domain datasets while mitigating images' artifacts. To validate the effectiveness of domain-invariant representations, we employ five widely used machine learning classifiers to construct blood cell classification models. Experimental results on two public blood cell datasets and a private real dataset demonstrate that our proposed DoRL achieves a new state-of-the-art cross-domain performance, surpassing existing methods by a significant margin. The source code can be available at the URL (https://github.com/AnoK3111/DoRL).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge