Xiangzhong Zhang
Stain-aware Domain Alignment for Imbalance Blood Cell Classification
Dec 04, 2024



Abstract:Blood cell identification is critical for hematological analysis as it aids physicians in diagnosing various blood-related diseases. In real-world scenarios, blood cell image datasets often present the issues of domain shift and data imbalance, posing challenges for accurate blood cell identification. To address these issues, we propose a novel blood cell classification method termed SADA via stain-aware domain alignment. The primary objective of this work is to mine domain-invariant features in the presence of domain shifts and data imbalances. To accomplish this objective, we propose a stain-based augmentation approach and a local alignment constraint to learn domain-invariant features. Furthermore, we propose a domain-invariant supervised contrastive learning strategy to capture discriminative features. We decouple the training process into two stages of domain-invariant feature learning and classification training, alleviating the problem of data imbalance. Experiment results on four public blood cell datasets and a private real dataset collected from the Third Affiliated Hospital of Sun Yat-sen University demonstrate that SADA can achieve a new state-of-the-art baseline, which is superior to the existing cutting-edge methods with a big margin. The source code can be available at the URL (\url{https://github.com/AnoK3111/SADA}).
Domain-invariant Representation Learning via Segment Anything Model for Blood Cell Classification
Aug 14, 2024Abstract:Accurate classification of blood cells is of vital significance in the diagnosis of hematological disorders. However, in real-world scenarios, domain shifts caused by the variability in laboratory procedures and settings, result in a rapid deterioration of the model's generalization performance. To address this issue, we propose a novel framework of domain-invariant representation learning (DoRL) via segment anything model (SAM) for blood cell classification. The DoRL comprises two main components: a LoRA-based SAM (LoRA-SAM) and a cross-domain autoencoder (CAE). The advantage of DoRL is that it can extract domain-invariant representations from various blood cell datasets in an unsupervised manner. Specifically, we first leverage the large-scale foundation model of SAM, fine-tuned with LoRA, to learn general image embeddings and segment blood cells. Additionally, we introduce CAE to learn domain-invariant representations across different-domain datasets while mitigating images' artifacts. To validate the effectiveness of domain-invariant representations, we employ five widely used machine learning classifiers to construct blood cell classification models. Experimental results on two public blood cell datasets and a private real dataset demonstrate that our proposed DoRL achieves a new state-of-the-art cross-domain performance, surpassing existing methods by a significant margin. The source code can be available at the URL (https://github.com/AnoK3111/DoRL).
Towards Cross-Domain Single Blood Cell Image Classification via Large-Scale LoRA-based Segment Anything Model
Aug 13, 2024
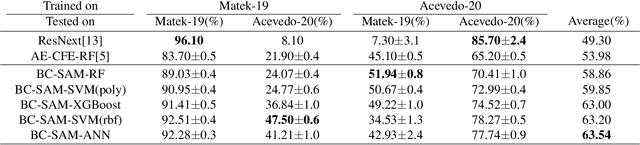
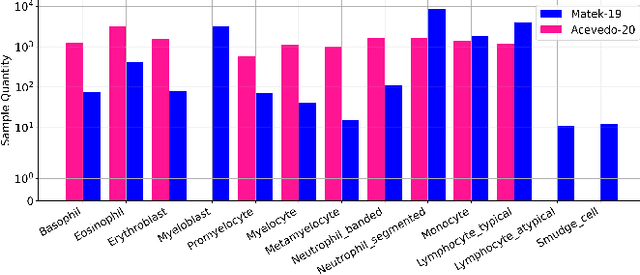
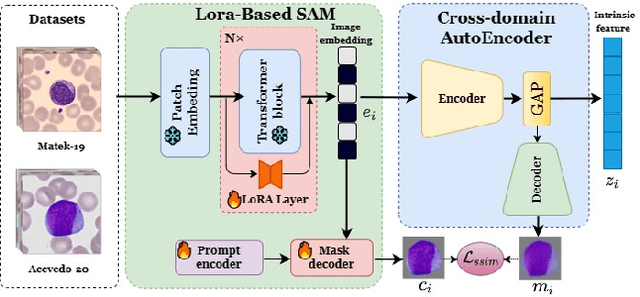
Abstract:Accurate classification of blood cells plays a vital role in hematological analysis as it aids physicians in diagnosing various medical conditions. In this study, we present a novel approach for classifying blood cell images known as BC-SAM. BC-SAM leverages the large-scale foundation model of Segment Anything Model (SAM) and incorporates a fine-tuning technique using LoRA, allowing it to extract general image embeddings from blood cell images. To enhance the applicability of BC-SAM across different blood cell image datasets, we introduce an unsupervised cross-domain autoencoder that focuses on learning intrinsic features while suppressing artifacts in the images. To assess the performance of BC-SAM, we employ four widely used machine learning classifiers (Random Forest, Support Vector Machine, Artificial Neural Network, and XGBoost) to construct blood cell classification models and compare them against existing state-of-the-art methods. Experimental results conducted on two publicly available blood cell datasets (Matek-19 and Acevedo-20) demonstrate that our proposed BC-SAM achieves a new state-of-the-art result, surpassing the baseline methods with a significant improvement. The source code of this paper is available at https://github.com/AnoK3111/BC-SAM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge