Genan Dai
ToPT: Task-Oriented Prompt Tuning for Urban Region Representation Learning
Feb 02, 2026Abstract:Learning effective region embeddings from heterogeneous urban data underpins key urban computing tasks (e.g., crime prediction, resource allocation). However, prevailing two-stage methods yield task-agnostic representations, decoupling them from downstream objectives. Recent prompt-based approaches attempt to fix this but introduce two challenges: they often lack explicit spatial priors, causing spatially incoherent inter-region modeling, and they lack robust mechanisms for explicit task-semantic alignment. We propose ToPT, a two-stage framework that delivers spatially consistent fusion and explicit task alignment. ToPT consists of two modules: spatial-aware region embedding learning (SREL) and task-aware prompting for region embeddings (Prompt4RE). SREL employs a Graphormer-based fusion module that injects spatial priors-distance and regional centrality-as learnable attention biases to capture coherent, interpretable inter-region interactions. Prompt4RE performs task-oriented prompting: a frozen multimodal large language model (MLLM) processes task-specific templates to obtain semantic vectors, which are aligned with region embeddings via multi-head cross-attention for stable task conditioning. Experiments across multiple tasks and cities show state-of-the-art performance, with improvements of up to 64.2\%, validating the necessity and complementarity of spatial priors and prompt-region alignment. The code is available at https://github.com/townSeven/Prompt4RE.git.
Abstract, Align, Predict: Zero-Shot Stance Detection via Cognitive Inductive Reasoning
Jun 16, 2025Abstract:Zero-shot stance detection (ZSSD) aims to identify the stance of text toward previously unseen targets, a setting where conventional supervised models often fail due to reliance on labeled data and shallow lexical cues. Inspired by human cognitive reasoning, we propose the Cognitive Inductive Reasoning Framework (CIRF), which abstracts transferable reasoning schemas from unlabeled text and encodes them as concept-level logic. To integrate these schemas with input arguments, we introduce a Schema-Enhanced Graph Kernel Model (SEGKM) that dynamically aligns local and global reasoning structures. Experiments on SemEval-2016, VAST, and COVID-19-Stance benchmarks show that CIRF establishes new state-of-the-art results, outperforming strong ZSSD baselines by 1.0, 4.5, and 3.3 percentage points in macro-F1, respectively, and achieving comparable accuracy with 70\% fewer labeled examples. We will release the full code upon publication.
C-MTCSD: A Chinese Multi-Turn Conversational Stance Detection Dataset
Apr 14, 2025Abstract:Stance detection has become an essential tool for analyzing public discussions on social media. Current methods face significant challenges, particularly in Chinese language processing and multi-turn conversational analysis. To address these limitations, we introduce C-MTCSD, the largest Chinese multi-turn conversational stance detection dataset, comprising 24,264 carefully annotated instances from Sina Weibo, which is 4.2 times larger than the only prior Chinese conversational stance detection dataset. Our comprehensive evaluation using both traditional approaches and large language models reveals the complexity of C-MTCSD: even state-of-the-art models achieve only 64.07% F1 score in the challenging zero-shot setting, while performance consistently degrades with increasing conversation depth. Traditional models particularly struggle with implicit stance detection, achieving below 50% F1 score. This work establishes a challenging new benchmark for Chinese stance detection research, highlighting significant opportunities for future improvements.
Multimodal Multi-turn Conversation Stance Detection: A Challenge Dataset and Effective Model
Sep 01, 2024



Abstract:Stance detection, which aims to identify public opinion towards specific targets using social media data, is an important yet challenging task. With the proliferation of diverse multimodal social media content including text, and images multimodal stance detection (MSD) has become a crucial research area. However, existing MSD studies have focused on modeling stance within individual text-image pairs, overlooking the multi-party conversational contexts that naturally occur on social media. This limitation stems from a lack of datasets that authentically capture such conversational scenarios, hindering progress in conversational MSD. To address this, we introduce a new multimodal multi-turn conversational stance detection dataset (called MmMtCSD). To derive stances from this challenging dataset, we propose a novel multimodal large language model stance detection framework (MLLM-SD), that learns joint stance representations from textual and visual modalities. Experiments on MmMtCSD show state-of-the-art performance of our proposed MLLM-SD approach for multimodal stance detection. We believe that MmMtCSD will contribute to advancing real-world applications of stance detection research.
Domain-invariant Representation Learning via Segment Anything Model for Blood Cell Classification
Aug 14, 2024Abstract:Accurate classification of blood cells is of vital significance in the diagnosis of hematological disorders. However, in real-world scenarios, domain shifts caused by the variability in laboratory procedures and settings, result in a rapid deterioration of the model's generalization performance. To address this issue, we propose a novel framework of domain-invariant representation learning (DoRL) via segment anything model (SAM) for blood cell classification. The DoRL comprises two main components: a LoRA-based SAM (LoRA-SAM) and a cross-domain autoencoder (CAE). The advantage of DoRL is that it can extract domain-invariant representations from various blood cell datasets in an unsupervised manner. Specifically, we first leverage the large-scale foundation model of SAM, fine-tuned with LoRA, to learn general image embeddings and segment blood cells. Additionally, we introduce CAE to learn domain-invariant representations across different-domain datasets while mitigating images' artifacts. To validate the effectiveness of domain-invariant representations, we employ five widely used machine learning classifiers to construct blood cell classification models. Experimental results on two public blood cell datasets and a private real dataset demonstrate that our proposed DoRL achieves a new state-of-the-art cross-domain performance, surpassing existing methods by a significant margin. The source code can be available at the URL (https://github.com/AnoK3111/DoRL).
Towards Cross-Domain Single Blood Cell Image Classification via Large-Scale LoRA-based Segment Anything Model
Aug 13, 2024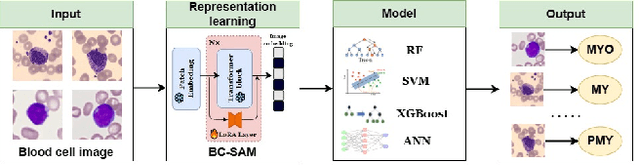
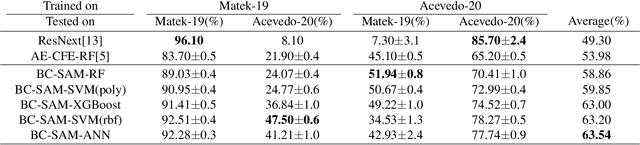
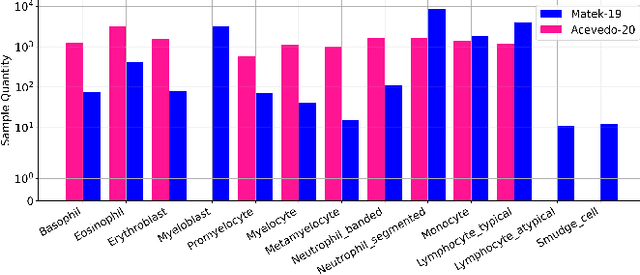
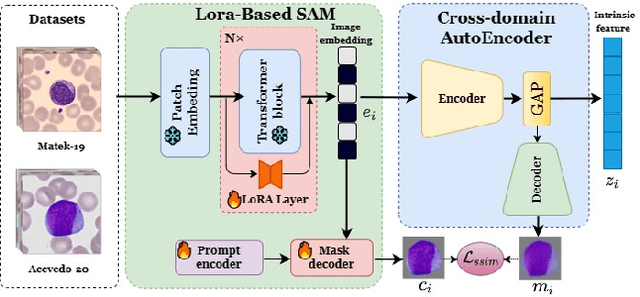
Abstract:Accurate classification of blood cells plays a vital role in hematological analysis as it aids physicians in diagnosing various medical conditions. In this study, we present a novel approach for classifying blood cell images known as BC-SAM. BC-SAM leverages the large-scale foundation model of Segment Anything Model (SAM) and incorporates a fine-tuning technique using LoRA, allowing it to extract general image embeddings from blood cell images. To enhance the applicability of BC-SAM across different blood cell image datasets, we introduce an unsupervised cross-domain autoencoder that focuses on learning intrinsic features while suppressing artifacts in the images. To assess the performance of BC-SAM, we employ four widely used machine learning classifiers (Random Forest, Support Vector Machine, Artificial Neural Network, and XGBoost) to construct blood cell classification models and compare them against existing state-of-the-art methods. Experimental results conducted on two publicly available blood cell datasets (Matek-19 and Acevedo-20) demonstrate that our proposed BC-SAM achieves a new state-of-the-art result, surpassing the baseline methods with a significant improvement. The source code of this paper is available at https://github.com/AnoK3111/BC-SAM.
Core Knowledge Learning Framework for Graph Adaptation and Scalability Learning
Jul 02, 2024



Abstract:Graph classification is a pivotal challenge in machine learning, especially within the realm of graph-based data, given its importance in numerous real-world applications such as social network analysis, recommendation systems, and bioinformatics. Despite its significance, graph classification faces several hurdles, including adapting to diverse prediction tasks, training across multiple target domains, and handling small-sample prediction scenarios. Current methods often tackle these challenges individually, leading to fragmented solutions that lack a holistic approach to the overarching problem. In this paper, we propose an algorithm aimed at addressing the aforementioned challenges. By incorporating insights from various types of tasks, our method aims to enhance adaptability, scalability, and generalizability in graph classification. Motivated by the recognition that the underlying subgraph plays a crucial role in GNN prediction, while the remainder is task-irrelevant, we introduce the Core Knowledge Learning (\method{}) framework for graph adaptation and scalability learning. \method{} comprises several key modules, including the core subgraph knowledge submodule, graph domain adaptation module, and few-shot learning module for downstream tasks. Each module is tailored to tackle specific challenges in graph classification, such as domain shift, label inconsistencies, and data scarcity. By learning the core subgraph of the entire graph, we focus on the most pertinent features for task relevance. Consequently, our method offers benefits such as improved model performance, increased domain adaptability, and enhanced robustness to domain variations. Experimental results demonstrate significant performance enhancements achieved by our method compared to state-of-the-art approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge