Jia Wei
Beijing Institute of Technology, China
Prior Knowledge-enhanced Spatio-temporal Epidemic Forecasting
Feb 25, 2026Abstract:Spatio-temporal epidemic forecasting is critical for public health management, yet existing methods often struggle with insensitivity to weak epidemic signals, over-simplified spatial relations, and unstable parameter estimation. To address these challenges, we propose the Spatio-Temporal priOr-aware Epidemic Predictor (STOEP), a novel hybrid framework that integrates implicit spatio-temporal priors and explicit expert priors. STOEP consists of three key components: (1) Case-aware Adjacency Learning (CAL), which dynamically adjusts mobility-based regional dependencies using historical infection patterns; (2) Space-informed Parameter Estimating (SPE), which employs learnable spatial priors to amplify weak epidemic signals; and (3) Filter-based Mechanistic Forecasting (FMF), which uses an expert-guided adaptive thresholding strategy to regularize epidemic parameters. Extensive experiments on real-world COVID-19 and influenza datasets demonstrate that STOEP outperforms the best baseline by 11.1% in RMSE. The system has been deployed at one provincial CDC in China to facilitate downstream applications.
ASGMamba: Adaptive Spectral Gating Mamba for Multivariate Time Series Forecasting
Feb 02, 2026Abstract:Long-term multivariate time series forecasting (LTSF) plays a crucial role in various high-performance computing applications, including real-time energy grid management and large-scale traffic flow simulation. However, existing solutions face a dilemma: Transformer-based models suffer from quadratic complexity, limiting their scalability on long sequences, while linear State Space Models (SSMs) often struggle to distinguish valuable signals from high-frequency noise, leading to wasted state capacity. To bridge this gap, we propose ASGMamba, an efficient forecasting framework designed for resource-constrained supercomputing environments. ASGMamba integrates a lightweight Adaptive Spectral Gating (ASG) mechanism that dynamically filters noise based on local spectral energy, enabling the Mamba backbone to focus its state evolution on robust temporal dynamics. Furthermore, we introduce a hierarchical multi-scale architecture with variable-specific Node Embeddings to capture diverse physical characteristics. Extensive experiments on nine benchmarks demonstrate that ASGMamba achieves state-of-the-art accuracy. While keeping strictly $$\mathcal{O}(L)$$ complexity we significantly reduce the memory usage on long-horizon tasks, thus establishing ASGMamba as a scalable solution for high-throughput forecasting in resource limited environments.The code is available at https://github.com/hit636/ASGMamba
AWEMixer: Adaptive Wavelet-Enhanced Mixer Network for Long-Term Time Series Forecasting
Nov 06, 2025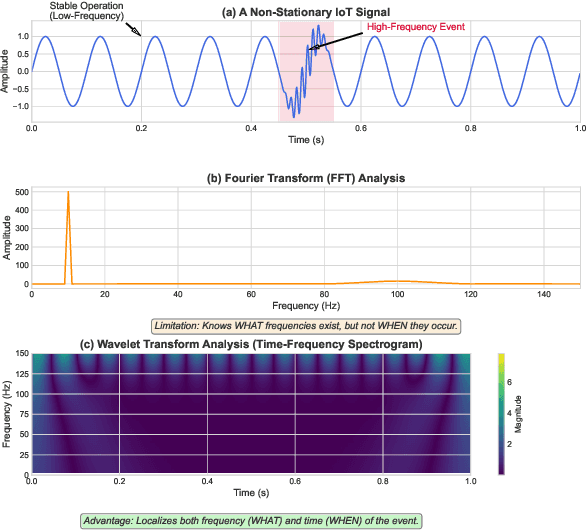
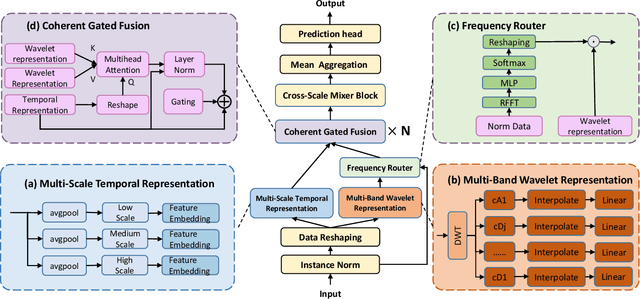
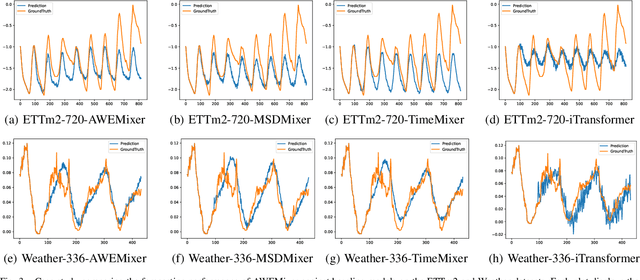
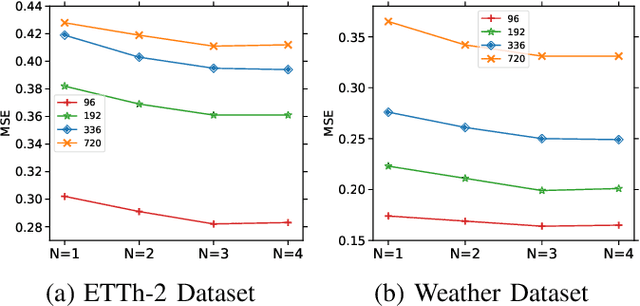
Abstract:Forecasting long-term time series in IoT environments remains a significant challenge due to the non-stationary and multi-scale characteristics of sensor signals. Furthermore, error accumulation causes a decrease in forecast quality when predicting further into the future. Traditional methods are restricted to operate in time-domain, while the global frequency information achieved by Fourier transform would be regarded as stationary signals leading to blur the temporal patterns of transient events. We propose AWEMixer, an Adaptive Wavelet-Enhanced Mixer Network including two innovative components: 1) a Frequency Router designs to utilize the global periodicity pattern achieved by Fast Fourier Transform to adaptively weight localized wavelet subband, and 2) a Coherent Gated Fusion Block to achieve selective integration of prominent frequency features with multi-scale temporal representation through cross-attention and gating mechanism, which realizes accurate time-frequency localization while remaining robust to noise. Seven public benchmarks validate that our model is more effective than recent state-of-the-art models. Specifically, our model consistently achieves performance improvement compared with transformer-based and MLP-based state-of-the-art models in long-sequence time series forecasting. Code is available at https://github.com/hit636/AWEMixer
DPANet: Dual Pyramid Attention Network for Multivariate Time Series Forecasting
Sep 18, 2025Abstract:We conducted rigorous ablation studies to validate DPANet's key components (Table \ref{tab:ablation-study}). The full model consistently outperforms all variants. To test our dual-domain hypothesis, we designed two specialized versions: a Temporal-Only model (fusing two identical temporal pyramids) and a Frequency-Only model (fusing two spectral pyramids). Both variants underperformed significantly, confirming that the fusion of heterogeneous temporal and frequency information is critical. Furthermore, replacing the cross-attention mechanism with a simpler method (w/o Cross-Fusion) caused the most severe performance degradation. This result underscores that our interactive fusion block is the most essential component.
SageAttention2++: A More Efficient Implementation of SageAttention2
May 28, 2025



Abstract:The efficiency of attention is critical because its time complexity grows quadratically with sequence length. SageAttention2 addresses this by utilizing quantization to accelerate matrix multiplications (Matmul) in attention. To further accelerate SageAttention2, we propose to utilize the faster instruction of FP8 Matmul accumulated in FP16. The instruction is 2x faster than the FP8 Matmul used in SageAttention2. Our experiments show that SageAttention2++ achieves a 3.9x speedup over FlashAttention while maintaining the same attention accuracy as SageAttention2. This means SageAttention2++ effectively accelerates various models, including those for language, image, and video generation, with negligible end-to-end metrics loss. The code will be available at https://github.com/thu-ml/SageAttention.
SageAttention3: Microscaling FP4 Attention for Inference and An Exploration of 8-Bit Training
May 16, 2025Abstract:The efficiency of attention is important due to its quadratic time complexity. We enhance the efficiency of attention through two key contributions: First, we leverage the new FP4 Tensor Cores in Blackwell GPUs to accelerate attention computation. Our implementation achieves 1038 TOPS on RTX5090, which is a 5x speedup over the fastest FlashAttention on RTX5090. Experiments show that our FP4 attention can accelerate inference of various models in a plug-and-play way. Second, we pioneer low-bit attention to training tasks. Existing low-bit attention works like FlashAttention3 and SageAttention focus only on inference. However, the efficiency of training large models is also important. To explore whether low-bit attention can be effectively applied to training tasks, we design an accurate and efficient 8-bit attention for both forward and backward propagation. Experiments indicate that 8-bit attention achieves lossless performance in fine-tuning tasks but exhibits slower convergence in pretraining tasks. The code will be available at https://github.com/thu-ml/SageAttention.
Mixture-of-Shape-Experts (MoSE): End-to-End Shape Dictionary Framework to Prompt SAM for Generalizable Medical Segmentation
Apr 13, 2025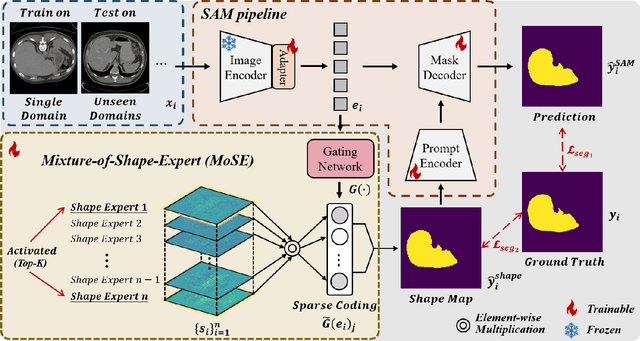
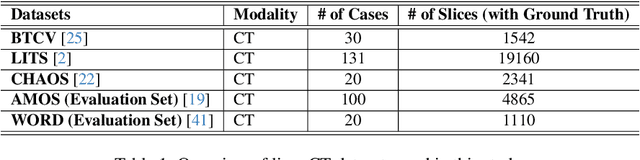
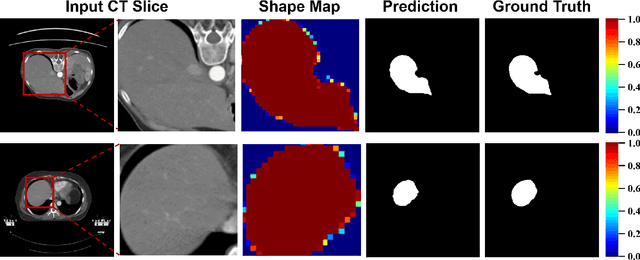
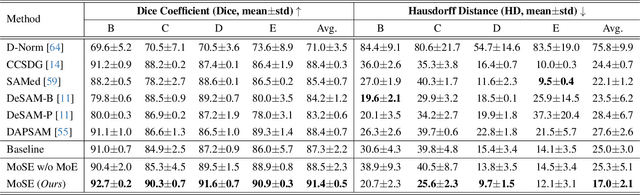
Abstract:Single domain generalization (SDG) has recently attracted growing attention in medical image segmentation. One promising strategy for SDG is to leverage consistent semantic shape priors across different imaging protocols, scanner vendors, and clinical sites. However, existing dictionary learning methods that encode shape priors often suffer from limited representational power with a small set of offline computed shape elements, or overfitting when the dictionary size grows. Moreover, they are not readily compatible with large foundation models such as the Segment Anything Model (SAM). In this paper, we propose a novel Mixture-of-Shape-Experts (MoSE) framework that seamlessly integrates the idea of mixture-of-experts (MoE) training into dictionary learning to efficiently capture diverse and robust shape priors. Our method conceptualizes each dictionary atom as a shape expert, which specializes in encoding distinct semantic shape information. A gating network dynamically fuses these shape experts into a robust shape map, with sparse activation guided by SAM encoding to prevent overfitting. We further provide this shape map as a prompt to SAM, utilizing the powerful generalization capability of SAM through bidirectional integration. All modules, including the shape dictionary, are trained in an end-to-end manner. Extensive experiments on multiple public datasets demonstrate its effectiveness.
Accurate INT8 Training Through Dynamic Block-Level Fallback
Mar 11, 2025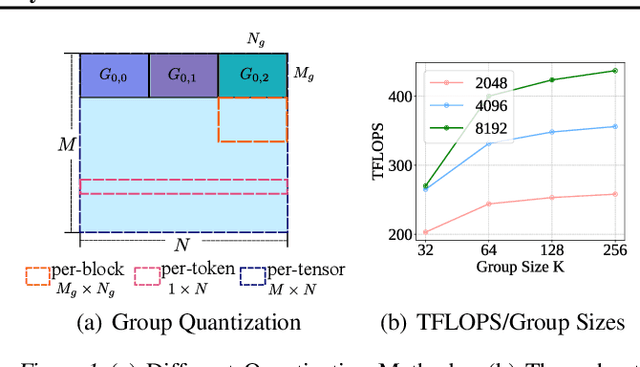
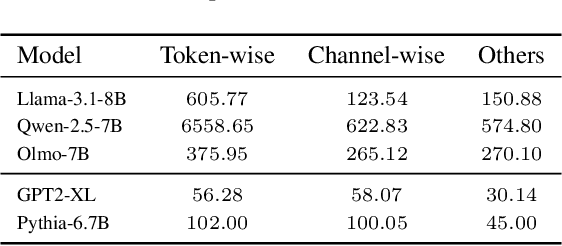
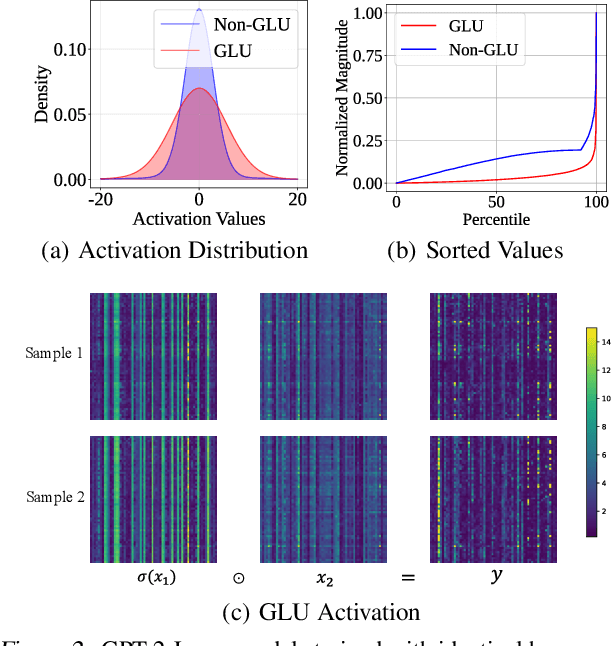
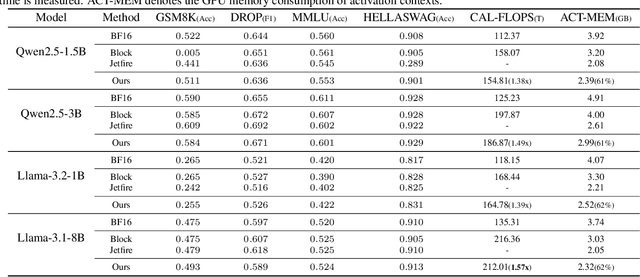
Abstract:Transformer models have achieved remarkable success across various AI applications but face significant training costs. Low-bit training, such as INT8 training, can leverage computational units with higher throughput, and has already demonstrated its effectiveness on GPT2 models with block-level quantization. However, it struggles with modern Transformer variants incorporating GLU units. This is because those variants demonstrate complex distributions of activation outliers. To address the challenge, we propose Fallback Quantization, implementing mixed-precision GEMM that dynamically falls back 8-bit to 16-bit for activation blocks containing outliers. Experiments show that our approach is robustly competent in both fine-tuning and pretraining settings. Moreover, our method achieves a 1.57x end-to-end training speedup on RTX4090 GPUs.
SpargeAttn: Accurate Sparse Attention Accelerating Any Model Inference
Feb 25, 2025Abstract:An efficient attention implementation is essential for large models due to its quadratic time complexity. Fortunately, attention commonly exhibits sparsity, i.e., many values in the attention map are near zero, allowing for the omission of corresponding computations. Many studies have utilized the sparse pattern to accelerate attention. However, most existing works focus on optimizing attention within specific models by exploiting certain sparse patterns of the attention map. A universal sparse attention that guarantees both the speedup and end-to-end performance of diverse models remains elusive. In this paper, we propose SpargeAttn, a universal sparse and quantized attention for any model. Our method uses a two-stage online filter: in the first stage, we rapidly and accurately predict the attention map, enabling the skip of some matrix multiplications in attention. In the second stage, we design an online softmax-aware filter that incurs no extra overhead and further skips some matrix multiplications. Experiments show that our method significantly accelerates diverse models, including language, image, and video generation, without sacrificing end-to-end metrics. The codes are available at https://github.com/thu-ml/SpargeAttn.
DeFusion: An Effective Decoupling Fusion Network for Multi-Modal Pregnancy Prediction
Jan 08, 2025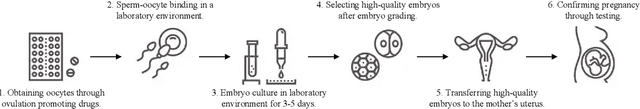
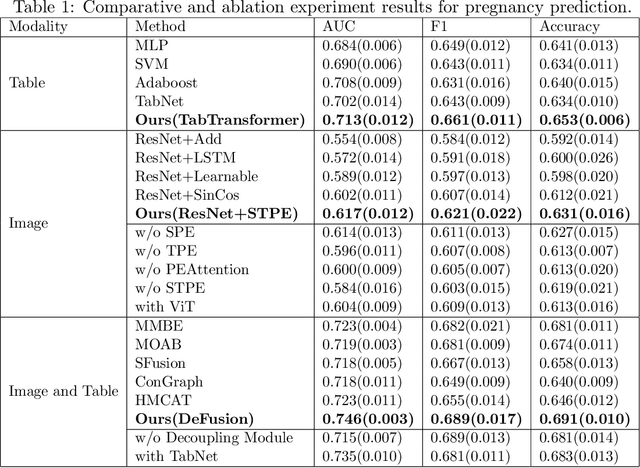
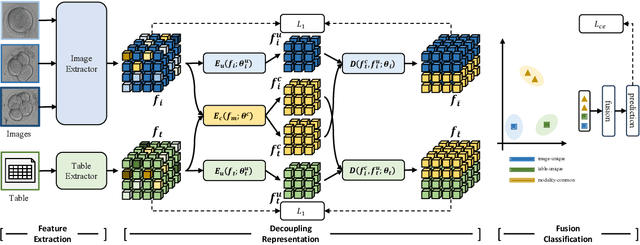
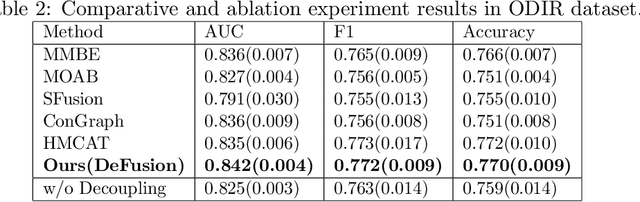
Abstract:Temporal embryo images and parental fertility table indicators are both valuable for pregnancy prediction in \textbf{in vitro fertilization embryo transfer} (IVF-ET). However, current machine learning models cannot make full use of the complementary information between the two modalities to improve pregnancy prediction performance. In this paper, we propose a Decoupling Fusion Network called DeFusion to effectively integrate the multi-modal information for IVF-ET pregnancy prediction. Specifically, we propose a decoupling fusion module that decouples the information from the different modalities into related and unrelated information, thereby achieving a more delicate fusion. And we fuse temporal embryo images with a spatial-temporal position encoding, and extract fertility table indicator information with a table transformer. To evaluate the effectiveness of our model, we use a new dataset including 4046 cases collected from Southern Medical University. The experiments show that our model outperforms state-of-the-art methods. Meanwhile, the performance on the eye disease prediction dataset reflects the model's good generalization. Our code and dataset are available at https://github.com/Ou-Young-1999/DFNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge