Jianyuan Deng
TRIDENT: Tri-Modal Molecular Representation Learning with Taxonomic Annotations and Local Correspondence
Jun 26, 2025Abstract:Molecular property prediction aims to learn representations that map chemical structures to functional properties. While multimodal learning has emerged as a powerful paradigm to learn molecular representations, prior works have largely overlooked textual and taxonomic information of molecules for representation learning. We introduce TRIDENT, a novel framework that integrates molecular SMILES, textual descriptions, and taxonomic functional annotations to learn rich molecular representations. To achieve this, we curate a comprehensive dataset of molecule-text pairs with structured, multi-level functional annotations. Instead of relying on conventional contrastive loss, TRIDENT employs a volume-based alignment objective to jointly align tri-modal features at the global level, enabling soft, geometry-aware alignment across modalities. Additionally, TRIDENT introduces a novel local alignment objective that captures detailed relationships between molecular substructures and their corresponding sub-textual descriptions. A momentum-based mechanism dynamically balances global and local alignment, enabling the model to learn both broad functional semantics and fine-grained structure-function mappings. TRIDENT achieves state-of-the-art performance on 11 downstream tasks, demonstrating the value of combining SMILES, textual, and taxonomic functional annotations for molecular property prediction.
Taking a Respite from Representation Learning for Molecular Property Prediction
Oct 07, 2022



Abstract:Artificial intelligence (AI) has been widely applied in drug discovery with a major task as molecular property prediction. Despite the boom of AI techniques in molecular representation learning, some key aspects underlying molecular property prediction haven't been carefully examined yet. In this study, we conducted a systematic comparison on three representative models, random forest, MolBERT and GROVER, which utilize three major molecular representations, extended-connectivity fingerprints, SMILES strings and molecular graphs, respectively. Notably, MolBERT and GROVER, are pretrained on large-scale unlabelled molecule corpuses in a self-supervised manner. In addition to the commonly used MoleculeNet benchmark datasets, we also assembled a suite of opioids-related datasets for downstream prediction evaluation. We first conducted dataset profiling on label distribution and structural analyses; we also examined the activity cliffs issue in the opioids-related datasets. Then, we trained 4,320 predictive models and evaluated the usefulness of the learned representations. Furthermore, we explored into the model evaluation by studying the effect of statistical tests, evaluation metrics and task settings. Finally, we dissected the chemical space generalization into inter-scaffold and intra-scaffold generalization and measured prediction performance to evaluate model generalizbility under both settings. By taking this respite, we reflected on the key aspects underlying molecular property prediction, the awareness of which can, hopefully, bring better AI techniques in this field.
Artificial Intelligence in Drug Discovery: Applications and Techniques
Jun 11, 2021



Abstract:Artificial intelligence (AI) has been transforming the practice of drug discovery in the past decade. Various AI techniques have been used in a wide range of applications, such as virtual screening and drug design. In this perspective, we first give an overview on drug discovery and discuss related applications, which can be reduced to two major tasks, i.e., molecular property prediction and molecule generation. We then discuss common data resources, molecule representations and benchmark platforms. Furthermore, to summarize the progress in AI-driven drug discovery, we present the relevant AI techniques including model architectures and learning paradigms in the surveyed papers. We expect that the perspective will serve as a guide for researchers who are interested in working at this intersected area of artificial intelligence and drug discovery. We also provide a GitHub repository\footnote{\url{https://github.com/dengjianyuan/Survey_AI_Drug_Discovery}} with the collection of papers and codes, if applicable, as a learning resource, which will be regularly updated.
Identifying Risk of Opioid Use Disorder for Patients Taking Opioid Medications with Deep Learning
Oct 09, 2020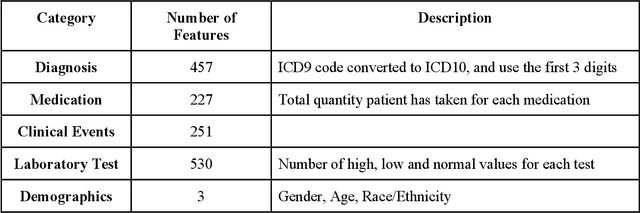
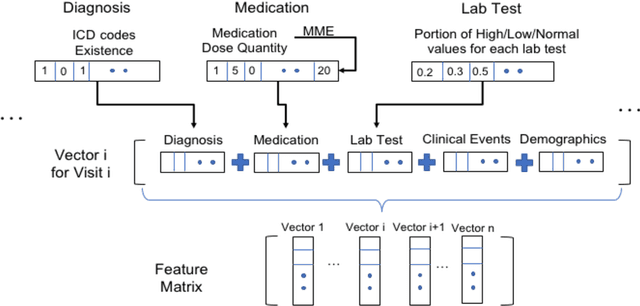
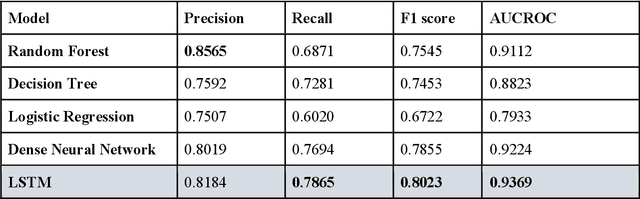
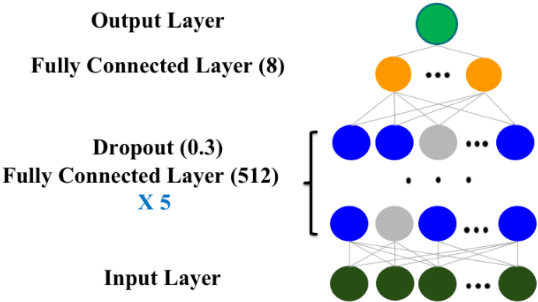
Abstract:The United States is experiencing an opioid epidemic, and there were more than 10 million opioid misusers aged 12 or older each year. Identifying patients at high risk of Opioid Use Disorder (OUD) can help to make early clinical interventions to reduce the risk of OUD. Our goal is to predict OUD patients among opioid prescription users through analyzing electronic health records with machine learning and deep learning methods. This will help us to better understand the diagnoses of OUD, providing new insights on opioid epidemic. Electronic health records of patients who have been prescribed with medications containing active opioid ingredients were extracted from Cerner Health Facts database between January 1, 2008 and December 31, 2017. Long Short-Term Memory (LSTM) models were applied to predict opioid use disorder risk in the future based on recent five encounters, and compared to Logistic Regression, Random Forest, Decision Tree and Dense Neural Network. Prediction performance was assessed using F-1 score, precision, recall, and AUROC. Our temporal deep learning model provided promising prediction results which outperformed other methods, with a F1 score of 0.8023 and AUCROC of 0.9369. The model can identify OUD related medications and vital signs as important features for the prediction. LSTM based temporal deep learning model is effective on predicting opioid use disorder using a patient past history of electronic health records, with minimal domain knowledge. It has potential to improve clinical decision support for early intervention and prevention to combat the opioid epidemic.
Towards Better Opioid Antagonists Using Deep Reinforcement Learning
Mar 26, 2020



Abstract:Naloxone, an opioid antagonist, has been widely used to save lives from opioid overdose, a leading cause for death in the opioid epidemic. However, naloxone has short brain retention ability, which limits its therapeutic efficacy. Developing better opioid antagonists is critical in combating the opioid epidemic.Instead of exhaustively searching in a huge chemical space for better opioid antagonists, we adopt reinforcement learning which allows efficient gradient-based search towards molecules with desired physicochemical and/or biological properties. Specifically, we implement a deep reinforcement learning framework to discover potential lead compounds as better opioid antagonists with enhanced brain retention ability. A customized multi-objective reward function is designed to bias the generation towards molecules with both sufficient opioid antagonistic effect and enhanced brain retention ability. Thorough evaluation demonstrates that with this framework, we are able to identify valid, novel and feasible molecules with multiple desired properties, which has high potential in drug discovery.
Robust and Fast Decoding of High-Capacity Color QR Codes for Mobile Applications
May 19, 2018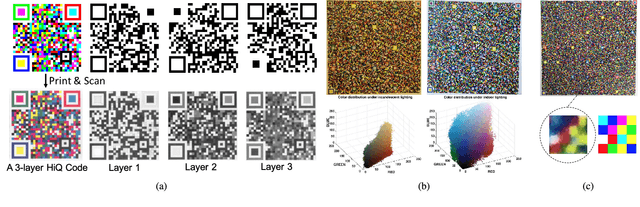
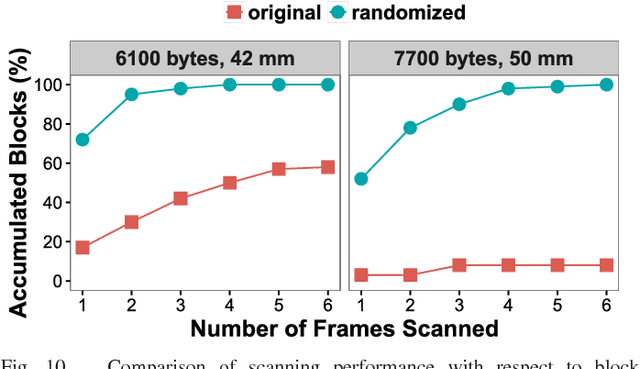


Abstract:The use of color in QR codes brings extra data capacity, but also inflicts tremendous challenges on the decoding process due to chromatic distortion, cross-channel color interference and illumination variation. Particularly, we further discover a new type of chromatic distortion in high-density color QR codes, cross-module color interference, caused by the high density which also makes the geometric distortion correction more challenging. To address these problems, we propose two approaches, namely, LSVM-CMI and QDA-CMI, which jointly model these different types of chromatic distortion. Extended from SVM and QDA, respectively, both LSVM-CMI and QDA-CMI optimize over a particular objective function to learn a color classifier. Furthermore, a robust geometric transformation method and several pipeline refinements are proposed to boost the decoding performance for mobile applications. We put forth and implement a framework for high-capacity color QR codes equipped with our methods, called HiQ. To evaluate the performance of HiQ, we collect a challenging large-scale color QR code dataset, CUHK-CQRC, which consists of 5390 high-density color QR code samples. The comparison with the baseline method [2] on CUHK-CQRC shows that HiQ at least outperforms [2] by 188% in decoding success rate and 60% in bit error rate. Our implementation of HiQ in iOS and Android also demonstrates the effectiveness of our framework in real-world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge