Fusheng Wang
BadCLM: Backdoor Attack in Clinical Language Models for Electronic Health Records
Jul 06, 2024Abstract:The advent of clinical language models integrated into electronic health records (EHR) for clinical decision support has marked a significant advancement, leveraging the depth of clinical notes for improved decision-making. Despite their success, the potential vulnerabilities of these models remain largely unexplored. This paper delves into the realm of backdoor attacks on clinical language models, introducing an innovative attention-based backdoor attack method, BadCLM (Bad Clinical Language Models). This technique clandestinely embeds a backdoor within the models, causing them to produce incorrect predictions when a pre-defined trigger is present in inputs, while functioning accurately otherwise. We demonstrate the efficacy of BadCLM through an in-hospital mortality prediction task with MIMIC III dataset, showcasing its potential to compromise model integrity. Our findings illuminate a significant security risk in clinical decision support systems and pave the way for future endeavors in fortifying clinical language models against such vulnerabilities.
Integrative Graph-Transformer Framework for Histopathology Whole Slide Image Representation and Classification
Mar 26, 2024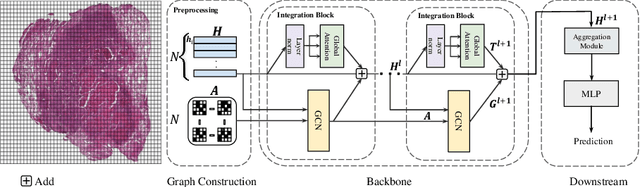
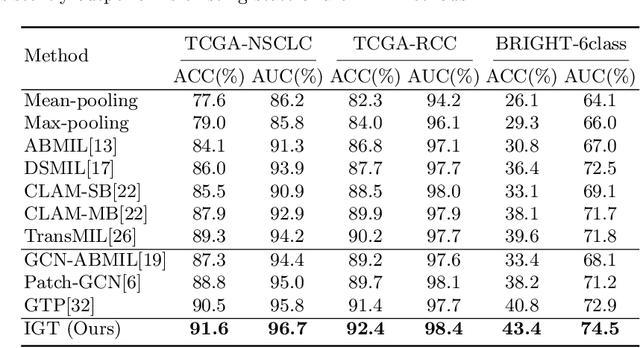
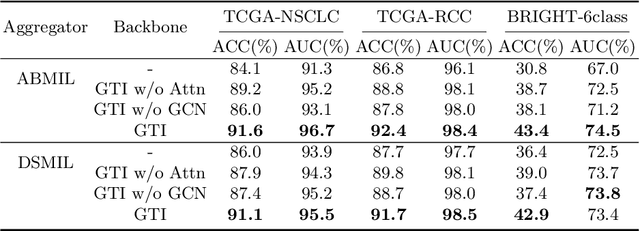
Abstract:In digital pathology, the multiple instance learning (MIL) strategy is widely used in the weakly supervised histopathology whole slide image (WSI) classification task where giga-pixel WSIs are only labeled at the slide level. However, existing attention-based MIL approaches often overlook contextual information and intrinsic spatial relationships between neighboring tissue tiles, while graph-based MIL frameworks have limited power to recognize the long-range dependencies. In this paper, we introduce the integrative graph-transformer framework that simultaneously captures the context-aware relational features and global WSI representations through a novel Graph Transformer Integration (GTI) block. Specifically, each GTI block consists of a Graph Convolutional Network (GCN) layer modeling neighboring relations at the local instance level and an efficient global attention model capturing comprehensive global information from extensive feature embeddings. Extensive experiments on three publicly available WSI datasets: TCGA-NSCLC, TCGA-RCC and BRIGHT, demonstrate the superiority of our approach over current state-of-the-art MIL methods, achieving an improvement of 1.0% to 2.6% in accuracy and 0.7%-1.6% in AUROC.
Enhancing Clinical Predictive Modeling through Model Complexity-Driven Class Proportion Tuning for Class Imbalanced Data: An Empirical Study on Opioid Overdose Prediction
May 09, 2023Abstract:Class imbalance problems widely exist in the medical field and heavily deteriorates performance of clinical predictive models. Most techniques to alleviate the problem rebalance class proportions and they predominantly assume the rebalanced proportions should be a function of the original data and oblivious to the model one uses. This work challenges this prevailing assumption and proposes that links the optimal class proportions to the model complexity, thereby tuning the class proportions per model. Our experiments on the opioid overdose prediction problem highlight the performance gain of tuning class proportions. Rigorous regression analysis also confirms the advantages of the theoretical framework proposed and the statistically significant correlation between the hyperparameters controlling the model complexity and the optimal class proportions.
Taking a Respite from Representation Learning for Molecular Property Prediction
Oct 07, 2022



Abstract:Artificial intelligence (AI) has been widely applied in drug discovery with a major task as molecular property prediction. Despite the boom of AI techniques in molecular representation learning, some key aspects underlying molecular property prediction haven't been carefully examined yet. In this study, we conducted a systematic comparison on three representative models, random forest, MolBERT and GROVER, which utilize three major molecular representations, extended-connectivity fingerprints, SMILES strings and molecular graphs, respectively. Notably, MolBERT and GROVER, are pretrained on large-scale unlabelled molecule corpuses in a self-supervised manner. In addition to the commonly used MoleculeNet benchmark datasets, we also assembled a suite of opioids-related datasets for downstream prediction evaluation. We first conducted dataset profiling on label distribution and structural analyses; we also examined the activity cliffs issue in the opioids-related datasets. Then, we trained 4,320 predictive models and evaluated the usefulness of the learned representations. Furthermore, we explored into the model evaluation by studying the effect of statistical tests, evaluation metrics and task settings. Finally, we dissected the chemical space generalization into inter-scaffold and intra-scaffold generalization and measured prediction performance to evaluate model generalizbility under both settings. By taking this respite, we reflected on the key aspects underlying molecular property prediction, the awareness of which can, hopefully, bring better AI techniques in this field.
A Multimodal Transformer: Fusing Clinical Notes with Structured EHR Data for Interpretable In-Hospital Mortality Prediction
Aug 09, 2022
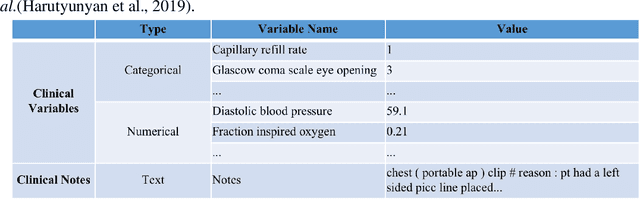
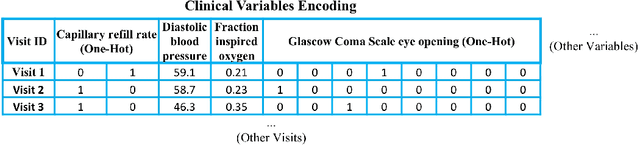
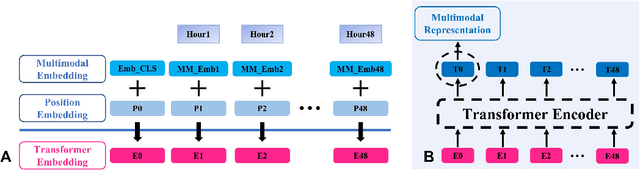
Abstract:Deep-learning-based clinical decision support using structured electronic health records (EHR) has been an active research area for predicting risks of mortality and diseases. Meanwhile, large amounts of narrative clinical notes provide complementary information, but are often not integrated into predictive models. In this paper, we provide a novel multimodal transformer to fuse clinical notes and structured EHR data for better prediction of in-hospital mortality. To improve interpretability, we propose an integrated gradients (IG) method to select important words in clinical notes and discover the critical structured EHR features with Shapley values. These important words and clinical features are visualized to assist with interpretation of the prediction outcomes. We also investigate the significance of domain adaptive pretraining and task adaptive fine-tuning on the Clinical BERT, which is used to learn the representations of clinical notes. Experiments demonstrated that our model outperforms other methods (AUCPR: 0.538, AUCROC: 0.877, F1:0.490).
Artificial Intelligence in Drug Discovery: Applications and Techniques
Jun 11, 2021



Abstract:Artificial intelligence (AI) has been transforming the practice of drug discovery in the past decade. Various AI techniques have been used in a wide range of applications, such as virtual screening and drug design. In this perspective, we first give an overview on drug discovery and discuss related applications, which can be reduced to two major tasks, i.e., molecular property prediction and molecule generation. We then discuss common data resources, molecule representations and benchmark platforms. Furthermore, to summarize the progress in AI-driven drug discovery, we present the relevant AI techniques including model architectures and learning paradigms in the surveyed papers. We expect that the perspective will serve as a guide for researchers who are interested in working at this intersected area of artificial intelligence and drug discovery. We also provide a GitHub repository\footnote{\url{https://github.com/dengjianyuan/Survey_AI_Drug_Discovery}} with the collection of papers and codes, if applicable, as a learning resource, which will be regularly updated.
Selective Knowledge Distillation for Neural Machine Translation
May 27, 2021



Abstract:Neural Machine Translation (NMT) models achieve state-of-the-art performance on many translation benchmarks. As an active research field in NMT, knowledge distillation is widely applied to enhance the model's performance by transferring teacher model's knowledge on each training sample. However, previous work rarely discusses the different impacts and connections among these samples, which serve as the medium for transferring teacher knowledge. In this paper, we design a novel protocol that can effectively analyze the different impacts of samples by comparing various samples' partitions. Based on above protocol, we conduct extensive experiments and find that the teacher's knowledge is not the more, the better. Knowledge over specific samples may even hurt the whole performance of knowledge distillation. Finally, to address these issues, we propose two simple yet effective strategies, i.e., batch-level and global-level selections, to pick suitable samples for distillation. We evaluate our approaches on two large-scale machine translation tasks, WMT'14 English->German and WMT'19 Chinese->English. Experimental results show that our approaches yield up to +1.28 and +0.89 BLEU points improvements over the Transformer baseline, respectively.
TRACE: Early Detection of Chronic Kidney Disease Onset with Transformer-Enhanced Feature Embedding
Dec 03, 2020
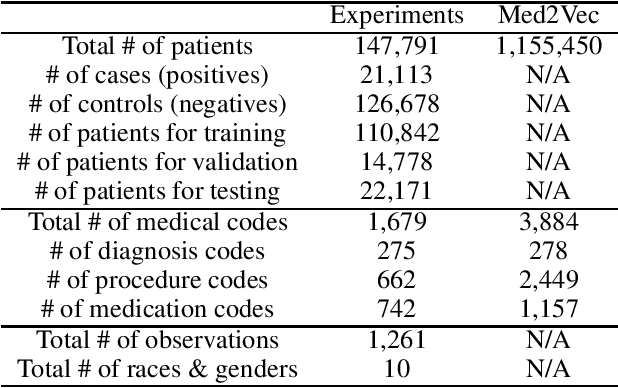
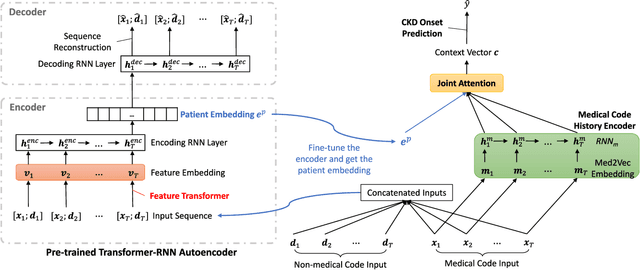
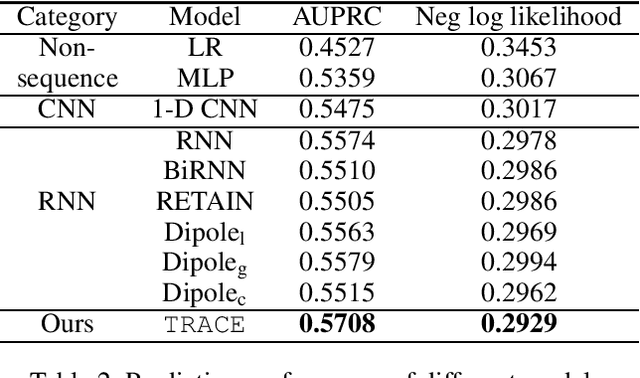
Abstract:Chronic kidney disease (CKD) has a poor prognosis due to excessive risk factors and comorbidities associated with it. The early detection of CKD faces challenges of insufficient medical histories of positive patients and complicated risk factors. In this paper, we propose the TRACE (Transformer-RNN Autoencoder-enhanced CKD Detector) framework, an end-to-end prediction model using patients' medical history data, to deal with these challenges. TRACE presents a comprehensive medical history representation with a novel key component: a Transformer-RNN autoencoder. The autoencoder jointly learns a medical concept embedding via Transformer for each hospital visit, and a latent representation which summarizes a patient's medical history across all the visits. We compared TRACE with multiple state-of-the-art methods on a dataset derived from real-world medical records. Our model has achieved 0.5708 AUPRC with a 2.31% relative improvement over the best-performing method. We also validated the clinical meaning of the learned embeddings through visualizations and a case study, showing the potential of TRACE to serve as a general disease prediction model.
Identifying Risk of Opioid Use Disorder for Patients Taking Opioid Medications with Deep Learning
Oct 09, 2020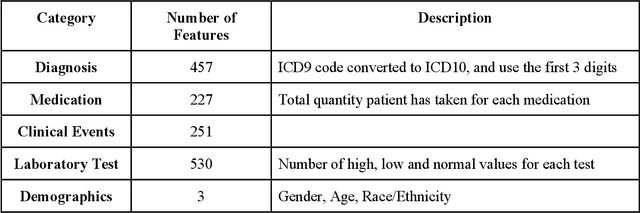
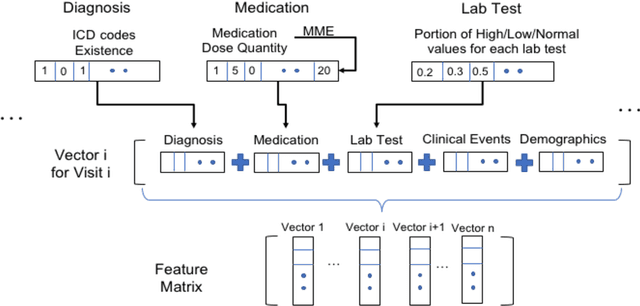
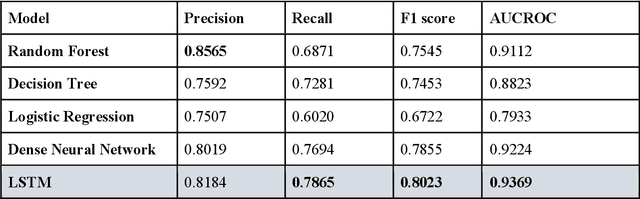
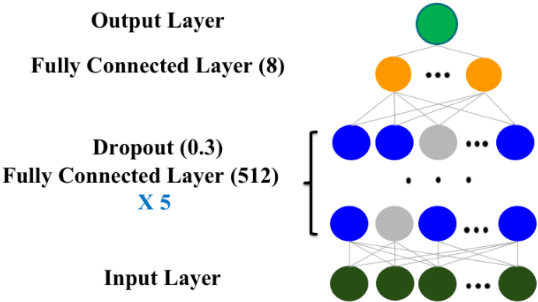
Abstract:The United States is experiencing an opioid epidemic, and there were more than 10 million opioid misusers aged 12 or older each year. Identifying patients at high risk of Opioid Use Disorder (OUD) can help to make early clinical interventions to reduce the risk of OUD. Our goal is to predict OUD patients among opioid prescription users through analyzing electronic health records with machine learning and deep learning methods. This will help us to better understand the diagnoses of OUD, providing new insights on opioid epidemic. Electronic health records of patients who have been prescribed with medications containing active opioid ingredients were extracted from Cerner Health Facts database between January 1, 2008 and December 31, 2017. Long Short-Term Memory (LSTM) models were applied to predict opioid use disorder risk in the future based on recent five encounters, and compared to Logistic Regression, Random Forest, Decision Tree and Dense Neural Network. Prediction performance was assessed using F-1 score, precision, recall, and AUROC. Our temporal deep learning model provided promising prediction results which outperformed other methods, with a F1 score of 0.8023 and AUCROC of 0.9369. The model can identify OUD related medications and vital signs as important features for the prediction. LSTM based temporal deep learning model is effective on predicting opioid use disorder using a patient past history of electronic health records, with minimal domain knowledge. It has potential to improve clinical decision support for early intervention and prevention to combat the opioid epidemic.
Towards Better Opioid Antagonists Using Deep Reinforcement Learning
Mar 26, 2020



Abstract:Naloxone, an opioid antagonist, has been widely used to save lives from opioid overdose, a leading cause for death in the opioid epidemic. However, naloxone has short brain retention ability, which limits its therapeutic efficacy. Developing better opioid antagonists is critical in combating the opioid epidemic.Instead of exhaustively searching in a huge chemical space for better opioid antagonists, we adopt reinforcement learning which allows efficient gradient-based search towards molecules with desired physicochemical and/or biological properties. Specifically, we implement a deep reinforcement learning framework to discover potential lead compounds as better opioid antagonists with enhanced brain retention ability. A customized multi-objective reward function is designed to bias the generation towards molecules with both sufficient opioid antagonistic effect and enhanced brain retention ability. Thorough evaluation demonstrates that with this framework, we are able to identify valid, novel and feasible molecules with multiple desired properties, which has high potential in drug discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge