Jun Kong
Sample-aware Adaptive Structured Pruning for Large Language Models
Mar 08, 2025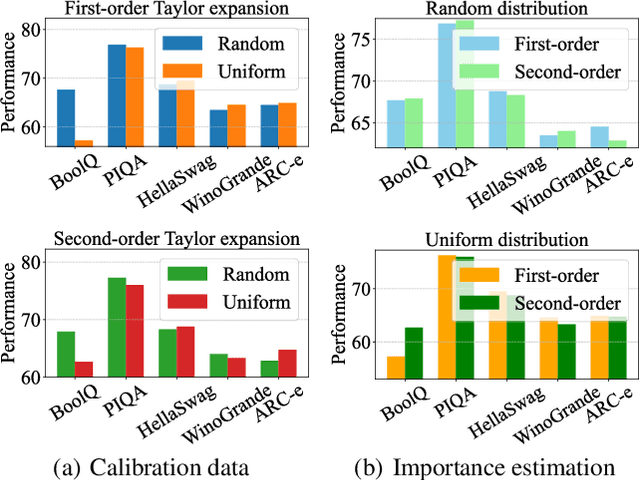
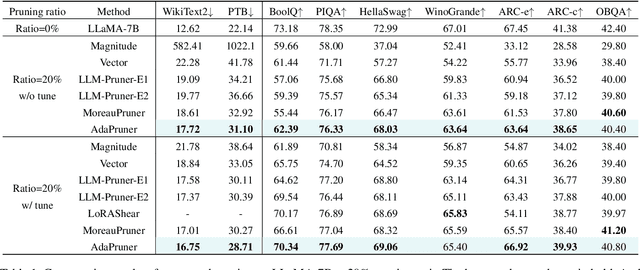
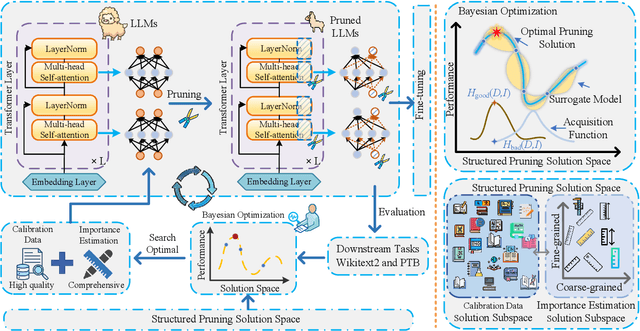
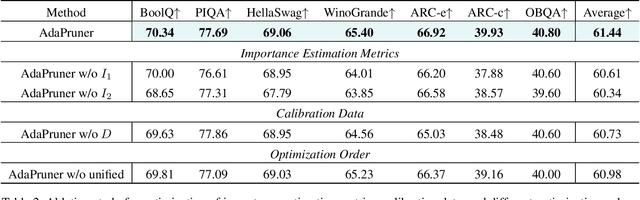
Abstract:Large language models (LLMs) have achieved outstanding performance in natural language processing, but enormous model sizes and high computational costs limit their practical deployment. Structured pruning can effectively reduce the resource demands for deployment by removing redundant model parameters. However, the randomly selected calibration data and fixed single importance estimation metrics in existing structured pruning methods lead to degraded performance of pruned models. This study introduces AdaPruner, a sample-aware adaptive structured pruning framework for LLMs, aiming to optimize the calibration data and importance estimation metrics in the structured pruning process. Specifically, AdaPruner effectively removes redundant parameters from LLMs by constructing a structured pruning solution space and then employing Bayesian optimization to adaptively search for the optimal calibration data and importance estimation metrics. Experimental results show that the AdaPruner outperforms existing structured pruning methods on a family of LLMs with varying pruning ratios, demonstrating its applicability and robustness. Remarkably, at a 20\% pruning ratio, the model pruned with AdaPruner maintains 97\% of the performance of the unpruned model.
Temporal Action Localization with Cross Layer Task Decoupling and Refinement
Dec 13, 2024Abstract:Temporal action localization (TAL) involves dual tasks to classify and localize actions within untrimmed videos. However, the two tasks often have conflicting requirements for features. Existing methods typically employ separate heads for classification and localization tasks but share the same input feature, leading to suboptimal performance. To address this issue, we propose a novel TAL method with Cross Layer Task Decoupling and Refinement (CLTDR). Based on the feature pyramid of video, CLTDR strategy integrates semantically strong features from higher pyramid layers and detailed boundary-aware boundary features from lower pyramid layers to effectively disentangle the action classification and localization tasks. Moreover, the multiple features from cross layers are also employed to refine and align the disentangled classification and regression results. At last, a lightweight Gated Multi-Granularity (GMG) module is proposed to comprehensively extract and aggregate video features at instant, local, and global temporal granularities. Benefiting from the CLTDR and GMG modules, our method achieves state-of-the-art performance on five challenging benchmarks: THUMOS14, MultiTHUMOS, EPIC-KITCHENS-100, ActivityNet-1.3, and HACS. Our code and pre-trained models are publicly available at: https://github.com/LiQiang0307/CLTDR-GMG.
NACNet: A Histology Context-aware Transformer Graph Convolution Network for Predicting Treatment Response to Neoadjuvant Chemotherapy in Triple Negative Breast Cancer
Nov 14, 2024



Abstract:Neoadjuvant chemotherapy (NAC) response prediction for triple negative breast cancer (TNBC) patients is a challenging task clinically as it requires understanding complex histology interactions within the tumor microenvironment (TME). Digital whole slide images (WSIs) capture detailed tissue information, but their giga-pixel size necessitates computational methods based on multiple instance learning, which typically analyze small, isolated image tiles without the spatial context of the TME. To address this limitation and incorporate TME spatial histology interactions in predicting NAC response for TNBC patients, we developed a histology context-aware transformer graph convolution network (NACNet). Our deep learning method identifies the histopathological labels on individual image tiles from WSIs, constructs a spatial TME graph, and represents each node with features derived from tissue texture and social network analysis. It predicts NAC response using a transformer graph convolution network model enhanced with graph isomorphism network layers. We evaluate our method with WSIs of a cohort of TNBC patient (N=105) and compared its performance with multiple state-of-the-art machine learning and deep learning models, including both graph and non-graph approaches. Our NACNet achieves 90.0% accuracy, 96.0% sensitivity, 88.0% specificity, and an AUC of 0.82, through eight-fold cross-validation, outperforming baseline models. These comprehensive experimental results suggest that NACNet holds strong potential for stratifying TNBC patients by NAC response, thereby helping to prevent overtreatment, improve patient quality of life, reduce treatment cost, and enhance clinical outcomes, marking an important advancement toward personalized breast cancer treatment.
Integrative Graph-Transformer Framework for Histopathology Whole Slide Image Representation and Classification
Mar 26, 2024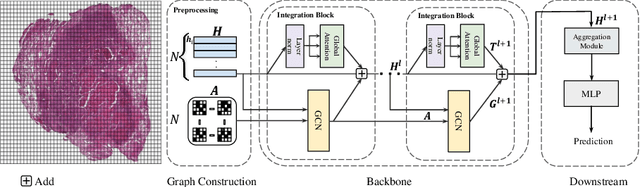
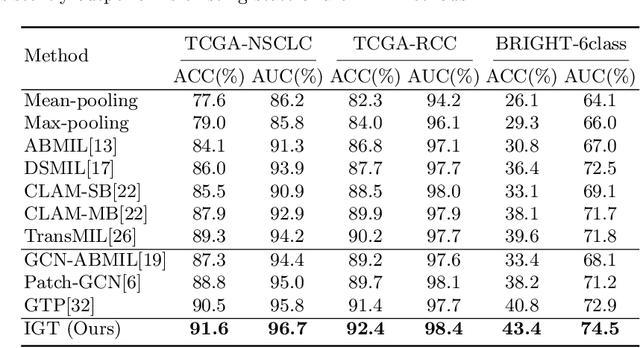
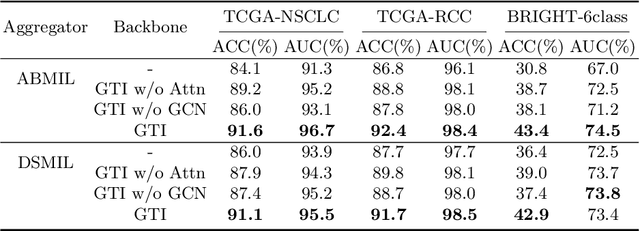
Abstract:In digital pathology, the multiple instance learning (MIL) strategy is widely used in the weakly supervised histopathology whole slide image (WSI) classification task where giga-pixel WSIs are only labeled at the slide level. However, existing attention-based MIL approaches often overlook contextual information and intrinsic spatial relationships between neighboring tissue tiles, while graph-based MIL frameworks have limited power to recognize the long-range dependencies. In this paper, we introduce the integrative graph-transformer framework that simultaneously captures the context-aware relational features and global WSI representations through a novel Graph Transformer Integration (GTI) block. Specifically, each GTI block consists of a Graph Convolutional Network (GCN) layer modeling neighboring relations at the local instance level and an efficient global attention model capturing comprehensive global information from extensive feature embeddings. Extensive experiments on three publicly available WSI datasets: TCGA-NSCLC, TCGA-RCC and BRIGHT, demonstrate the superiority of our approach over current state-of-the-art MIL methods, achieving an improvement of 1.0% to 2.6% in accuracy and 0.7%-1.6% in AUROC.
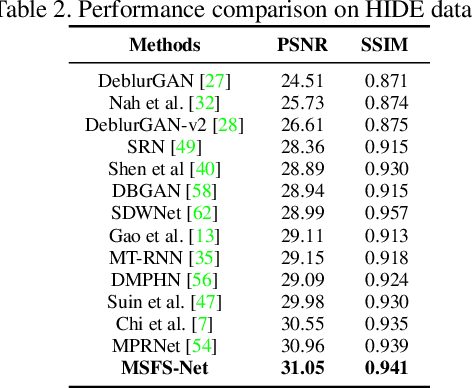
Multi-scale frequency separation network for image deblurring
Jun 01, 2022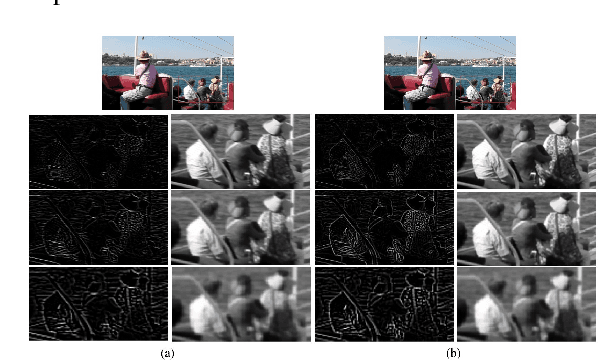

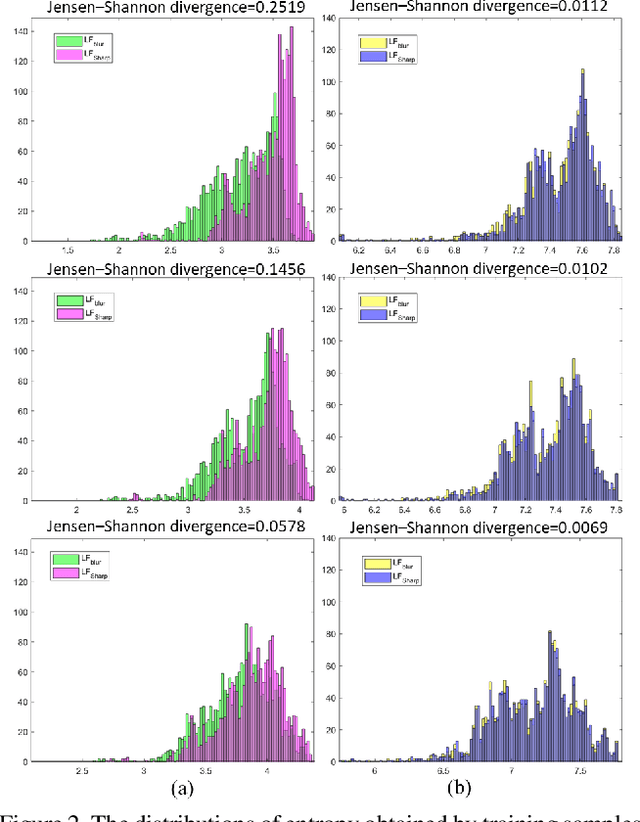
Abstract:Image deblurring aims to restore the detailed texture information or structures from the blurry images, which has become an indispensable step in many computer-vision tasks. Although various methods have been proposed to deal with the image deblurring problem, most of them treated the blurry image as a whole and neglected the characteristics of different image frequencies. In this paper, we present a new method called multi-scale frequency separation network (MSFS-Net) for image deblurring. MSFS-Net introduces the frequency separation module (FSM) into an encoder-decoder network architecture to capture the low and high-frequency information of image at multiple scales. Then, a simple cycle-consistency strategy and a sophisticated contrastive learning module (CLM) are respectively designed to retain the low-frequency information and recover the high-frequency information during deblurring. At last, the features of different scales are fused by a cross-scale feature fusion module (CSFFM). Extensive experiments on benchmark datasets show that the proposed network achieves state-of-the-art performance.
Last-iterate convergence analysis of stochastic momentum methods for neural networks
May 30, 2022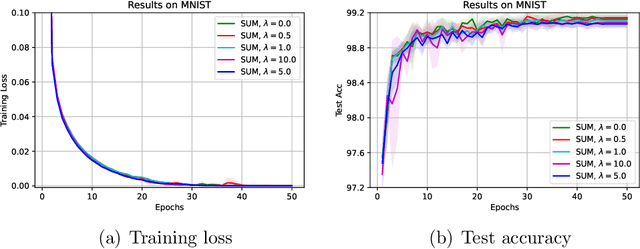
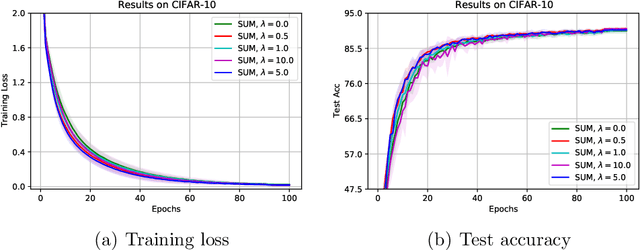
Abstract:The stochastic momentum method is a commonly used acceleration technique for solving large-scale stochastic optimization problems in artificial neural networks. Current convergence results of stochastic momentum methods under non-convex stochastic settings mostly discuss convergence in terms of the random output and minimum output. To this end, we address the convergence of the last iterate output (called last-iterate convergence) of the stochastic momentum methods for non-convex stochastic optimization problems, in a way conformal with traditional optimization theory. We prove the last-iterate convergence of the stochastic momentum methods under a unified framework, covering both stochastic heavy ball momentum and stochastic Nesterov accelerated gradient momentum. The momentum factors can be fixed to be constant, rather than time-varying coefficients in existing analyses. Finally, the last-iterate convergence of the stochastic momentum methods is verified on the benchmark MNIST and CIFAR-10 datasets.
Image deblurring based on lightweight multi-information fusion network
Jan 14, 2021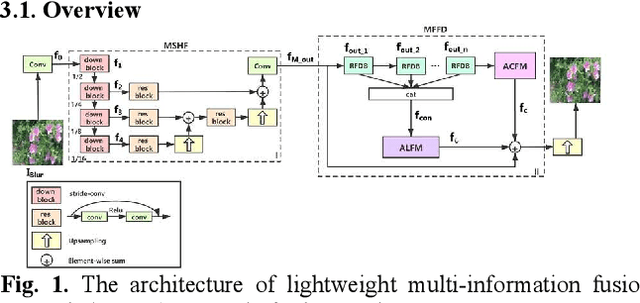
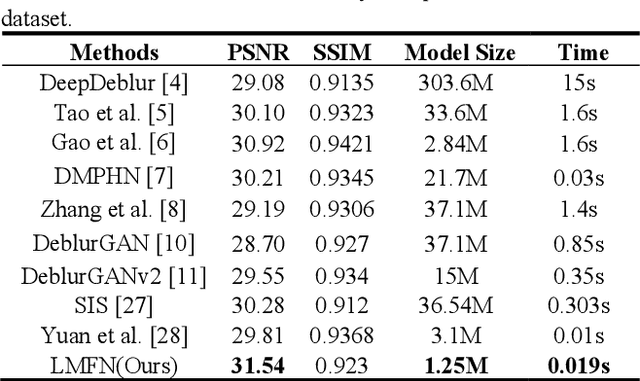
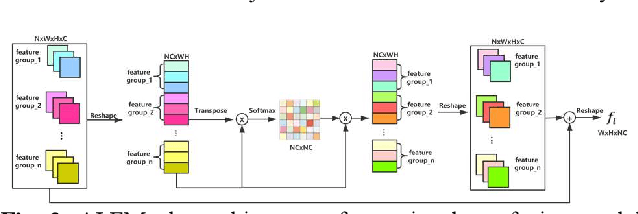

Abstract:Recently, deep learning based image deblurring has been well developed. However, exploiting the detailed image features in a deep learning framework always requires a mass of parameters, which inevitably makes the network suffer from high computational burden. To solve this problem, we propose a lightweight multiinformation fusion network (LMFN) for image deblurring. The proposed LMFN is designed as an encoder-decoder architecture. In the encoding stage, the image feature is reduced to various smallscale spaces for multi-scale information extraction and fusion without a large amount of information loss. Then, a distillation network is used in the decoding stage, which allows the network benefit the most from residual learning while remaining sufficiently lightweight. Meanwhile, an information fusion strategy between distillation modules and feature channels is also carried out by attention mechanism. Through fusing different information in the proposed approach, our network can achieve state-of-the-art image deblurring result with smaller number of parameters and outperforms existing methods in model complexity.
HPCC-YNU at SemEval-2020 Task 9: A Bilingual Vector Gating Mechanism for Sentiment Analysis of Code-Mixed Text
Oct 10, 2020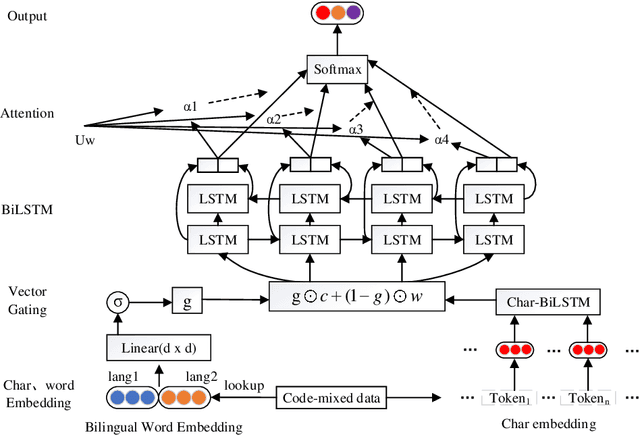
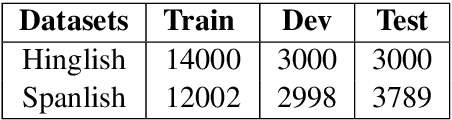
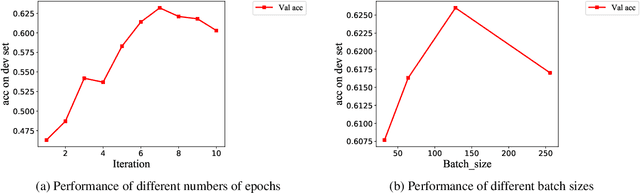

Abstract:It is fairly common to use code-mixing on a social media platform to express opinions and emotions in multilingual societies. The purpose of this task is to detect the sentiment of code-mixed social media text. Code-mixed text poses a great challenge for the traditional NLP system, which currently uses monolingual resources to deal with the problem of multilingual mixing. This task has been solved in the past using lexicon lookup in respective sentiment dictionaries and using a long short-term memory (LSTM) neural network for monolingual resources. In this paper, we (my codalab username is kongjun) present a system that uses a bilingual vector gating mechanism for bilingual resources to complete the task. The model consists of two main parts: the vector gating mechanism, which combines the character and word levels, and the attention mechanism, which extracts the important emotional parts of the text. The results show that the proposed system outperforms the baseline algorithm. We achieved fifth place in Spanglish and 19th place in Hinglish.The code of this paper is availabled at : https://github.com/JunKong5/Semveal2020-task9
Liver Steatosis Segmentation with Deep Learning Methods
Nov 16, 2019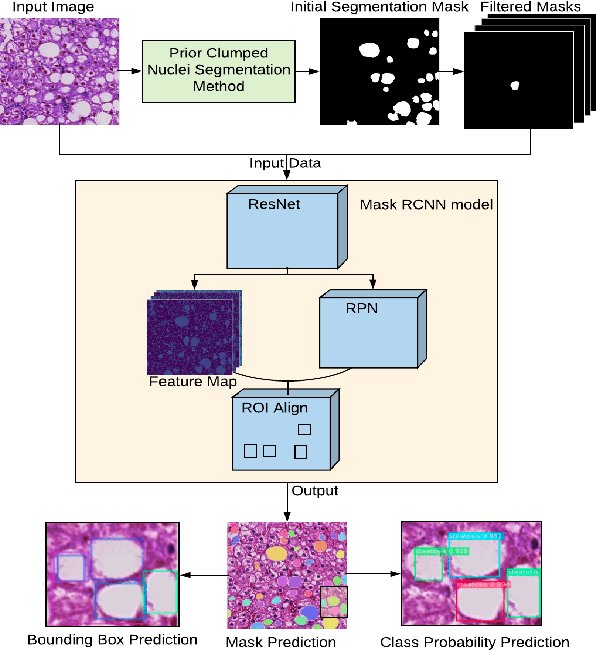
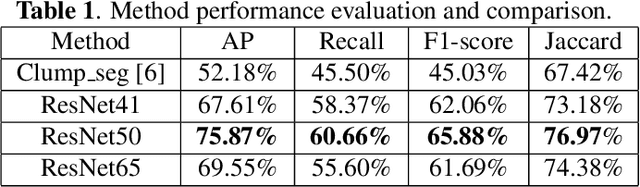
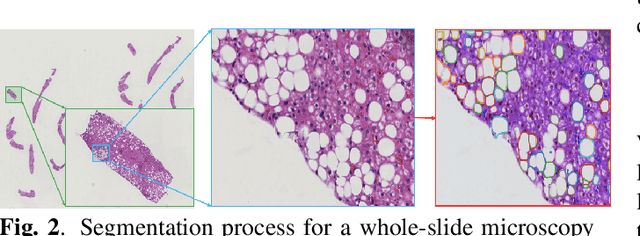

Abstract:Liver steatosis is known as the abnormal accumulation of lipids within cells. An accurate quantification of steatosis area within the liver histopathological microscopy images plays an important role in liver disease diagnosis and trans-plantation assessment. Such a quantification analysis often requires a precise steatosis segmentation that is challenging due to abundant presence of highly overlapped steatosis droplets. In this paper, a deep learning model Mask-RCNN is used to segment the steatosis droplets in clumps. Extended from Faster R-CNN, Mask-RCNN can predict object masks in addition to bounding box detection. With transfer learning, the resulting model is able to segment overlapped steatosis regions at 75.87% by Average Precision, 60.66% by Recall,65.88% by F1-score, and 76.97% by Jaccard index, promising to support liver disease diagnosis and allograft rejection prediction in future clinical practice.
* 4 pages
Clumped Nuclei Segmentation with Adjacent Point Match and Local Shape based Intensity Analysis for Overlapped Nuclei in Fluorescence In-Situ Hybridization Images
Aug 14, 2018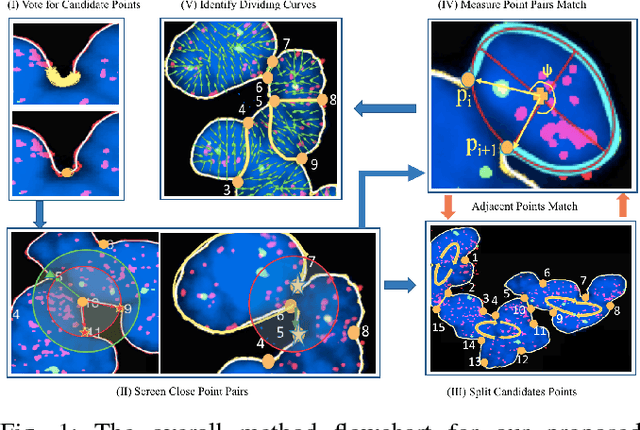
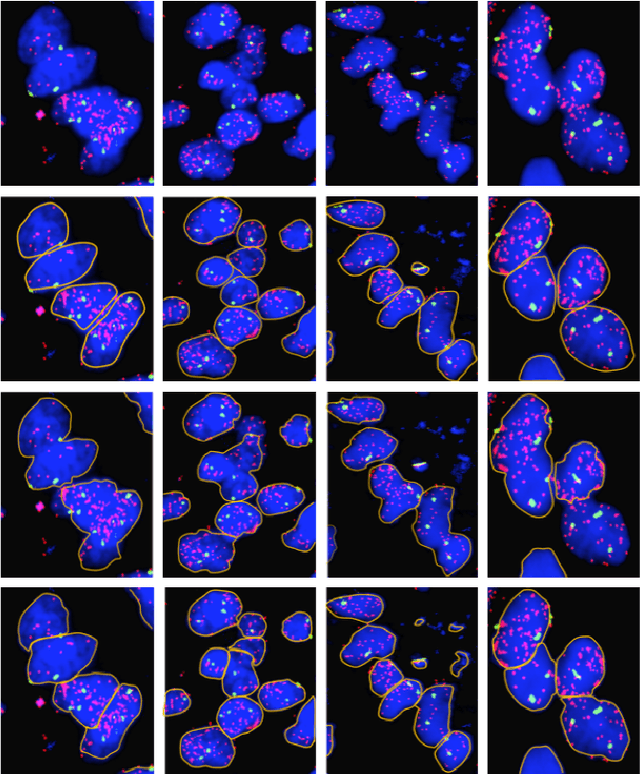
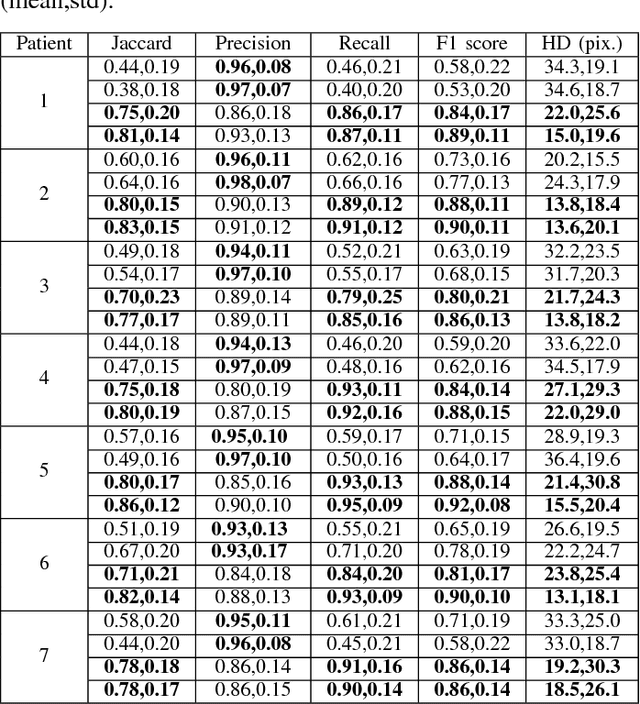
Abstract:Highly clumped nuclei clusters captured in fluorescence in situ hybridization microscopy images are common histology entities under investigations in a wide spectrum of tissue-related biomedical investigations. Due to their large scale in presence, computer based image analysis is used to facilitate such analysis with improved analysis efficiency and reproducibility. To ensure the quality of downstream biomedical analyses, it is essential to segment clustered nuclei with high quality. However, this presents a technical challenge commonly encountered in a large number of biomedical research, as nuclei are often overlapped due to a high cell density. In this paper, we propose an segmentation algorithm that identifies point pair connection candidates and evaluates adjacent point connections with a formulated ellipse fitting quality indicator. After connection relationships are determined, we recover the resulting dividing paths by following points with specific eigenvalues from Hessian in a constrained searching space. We validate our algorithm with 560 image patches from two classes of tumor regions of seven brain tumor patients. Both qualitative and quantitative experimental results suggest that our algorithm is promising for dividing overlapped nuclei in fluorescence in situ hybridization microscopy images widely used in various biomedical research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge