Manuel Schürch
Learning Personalized Treatment Decisions in Precision Medicine: Disentangling Treatment Assignment Bias in Counterfactual Outcome Prediction and Biomarker Identification
Oct 01, 2024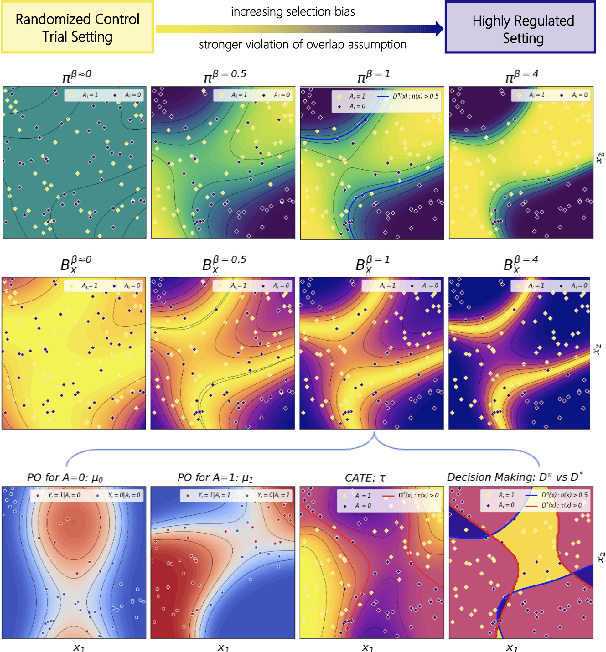
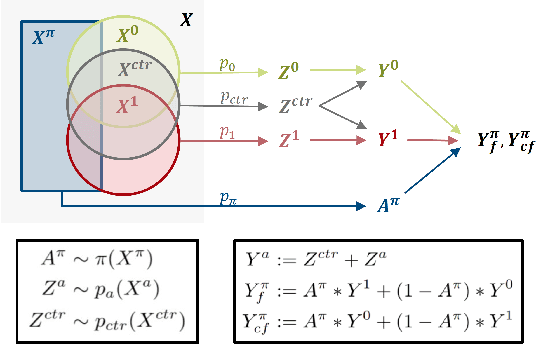
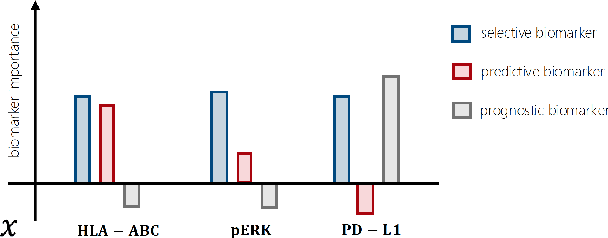
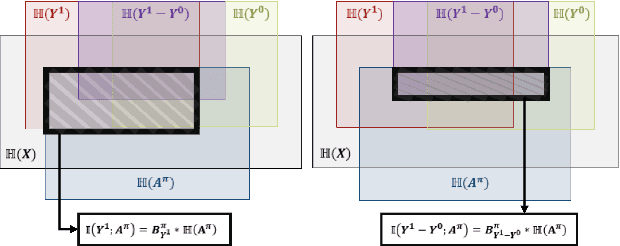
Abstract:Precision medicine offers the potential to tailor treatment decisions to individual patients, yet it faces significant challenges due to the complex biases in clinical observational data and the high-dimensional nature of biological data. This study models various types of treatment assignment biases using mutual information and investigates their impact on machine learning (ML) models for counterfactual prediction and biomarker identification. Unlike traditional counterfactual benchmarks that rely on fixed treatment policies, our work focuses on modeling different characteristics of the underlying observational treatment policy in distinct clinical settings. We validate our approach through experiments on toy datasets, semi-synthetic tumor cancer genome atlas (TCGA) data, and real-world biological outcomes from drug and CRISPR screens. By incorporating empirical biological mechanisms, we create a more realistic benchmark that reflects the complexities of real-world data. Our analysis reveals that different biases lead to varying model performances, with some biases, especially those unrelated to outcome mechanisms, having minimal effect on prediction accuracy. This highlights the crucial need to account for specific biases in clinical observational data in counterfactual ML model development, ultimately enhancing the personalization of treatment decisions in precision medicine.
Semi-Supervised Generative Models for Disease Trajectories: A Case Study on Systemic Sclerosis
Jul 16, 2024Abstract:We propose a deep generative approach using latent temporal processes for modeling and holistically analyzing complex disease trajectories, with a particular focus on Systemic Sclerosis (SSc). We aim to learn temporal latent representations of the underlying generative process that explain the observed patient disease trajectories in an interpretable and comprehensive way. To enhance the interpretability of these latent temporal processes, we develop a semi-supervised approach for disentangling the latent space using established medical knowledge. By combining the generative approach with medical definitions of different characteristics of SSc, we facilitate the discovery of new aspects of the disease. We show that the learned temporal latent processes can be utilized for further data analysis and clinical hypothesis testing, including finding similar patients and clustering SSc patient trajectories into novel sub-types. Moreover, our method enables personalized online monitoring and prediction of multivariate time series with uncertainty quantification.
Clustering of Disease Trajectories with Explainable Machine Learning: A Case Study on Postoperative Delirium Phenotypes
May 06, 2024



Abstract:The identification of phenotypes within complex diseases or syndromes is a fundamental component of precision medicine, which aims to adapt healthcare to individual patient characteristics. Postoperative delirium (POD) is a complex neuropsychiatric condition with significant heterogeneity in its clinical manifestations and underlying pathophysiology. We hypothesize that POD comprises several distinct phenotypes, which cannot be directly observed in clinical practice. Identifying these phenotypes could enhance our understanding of POD pathogenesis and facilitate the development of targeted prevention and treatment strategies. In this paper, we propose an approach that combines supervised machine learning for personalized POD risk prediction with unsupervised clustering techniques to uncover potential POD phenotypes. We first demonstrate our approach using synthetic data, where we simulate patient cohorts with predefined phenotypes based on distinct sets of informative features. We aim to mimic any clinical disease with our synthetic data generation method. By training a predictive model and applying SHAP, we show that clustering patients in the SHAP feature importance space successfully recovers the true underlying phenotypes, outperforming clustering in the raw feature space. We then present a case study using real-world data from a cohort of elderly surgical patients. The results showcase the utility of our approach in uncovering clinically relevant subtypes of complex disorders like POD, paving the way for more precise and personalized treatment strategies.
Towards AI-Based Precision Oncology: A Machine Learning Framework for Personalized Counterfactual Treatment Suggestions based on Multi-Omics Data
Feb 19, 2024



Abstract:AI-driven precision oncology has the transformative potential to reshape cancer treatment by leveraging the power of AI models to analyze the interaction between complex patient characteristics and their corresponding treatment outcomes. New technological platforms have facilitated the timely acquisition of multimodal data on tumor biology at an unprecedented resolution, such as single-cell multi-omics data, making this quality and quantity of data available for data-driven improved clinical decision-making. In this work, we propose a modular machine learning framework designed for personalized counterfactual cancer treatment suggestions based on an ensemble of machine learning experts trained on diverse multi-omics technologies. These specialized counterfactual experts per technology are consistently aggregated into a more powerful expert with superior performance and can provide both confidence and an explanation of its decision. The framework is tailored to address critical challenges inherent in data-driven cancer research, including the high-dimensional nature of the data, and the presence of treatment assignment bias in the retrospective observational data. The framework is showcased through comprehensive demonstrations using data from in-vitro and in-vivo treatment responses from a cohort of patients with ovarian cancer. Our method aims to empower clinicians with a reality-centric decision-support tool including probabilistic treatment suggestions with calibrated confidence and personalized explanations for tailoring treatment strategies to multi-omics characteristics of individual cancer patients.
Modeling Complex Disease Trajectories using Deep Generative Models with Semi-Supervised Latent Processes
Nov 17, 2023



Abstract:In this paper, we propose a deep generative time series approach using latent temporal processes for modeling and holistically analyzing complex disease trajectories. We aim to find meaningful temporal latent representations of an underlying generative process that explain the observed disease trajectories in an interpretable and comprehensive way. To enhance the interpretability of these latent temporal processes, we develop a semi-supervised approach for disentangling the latent space using established medical concepts. By combining the generative approach with medical knowledge, we leverage the ability to discover novel aspects of the disease while integrating medical concepts into the model. We show that the learned temporal latent processes can be utilized for further data analysis and clinical hypothesis testing, including finding similar patients and clustering the disease into new sub-types. Moreover, our method enables personalized online monitoring and prediction of multivariate time series including uncertainty quantification. We demonstrate the effectiveness of our approach in modeling systemic sclerosis, showcasing the potential of our machine learning model to capture complex disease trajectories and acquire new medical knowledge.
Dynamic Local Attention with Hierarchical Patching for Irregular Clinical Time Series
Nov 13, 2023



Abstract:Irregular multivariate time series data is prevalent in the clinical and healthcare domains. It is characterized by time-wise and feature-wise irregularities, making it challenging for machine learning methods to work with. To solve this, we introduce a new model architecture composed of two modules: (1) DLA, a Dynamic Local Attention mechanism that uses learnable queries and feature-specific local windows when computing the self-attention operation. This results in aggregating irregular time steps raw input within each window to a harmonized regular latent space representation while taking into account the different features' sampling rates. (2) A hierarchical MLP mixer that processes the output of DLA through multi-scale patching to leverage information at various scales for the downstream tasks. Our approach outperforms state-of-the-art methods on three real-world datasets, including the latest clinical MIMIC IV dataset.
Generating Personalized Insulin Treatments Strategies with Deep Conditional Generative Time Series Models
Sep 28, 2023



Abstract:We propose a novel framework that combines deep generative time series models with decision theory for generating personalized treatment strategies. It leverages historical patient trajectory data to jointly learn the generation of realistic personalized treatment and future outcome trajectories through deep generative time series models. In particular, our framework enables the generation of novel multivariate treatment strategies tailored to the personalized patient history and trained for optimal expected future outcomes based on conditional expected utility maximization. We demonstrate our framework by generating personalized insulin treatment strategies and blood glucose predictions for hospitalized diabetes patients, showcasing the potential of our approach for generating improved personalized treatment strategies. Keywords: deep generative model, probabilistic decision support, personalized treatment generation, insulin and blood glucose prediction
SimTS: Rethinking Contrastive Representation Learning for Time Series Forecasting
Mar 31, 2023



Abstract:Contrastive learning methods have shown an impressive ability to learn meaningful representations for image or time series classification. However, these methods are less effective for time series forecasting, as optimization of instance discrimination is not directly applicable to predicting the future state from the history context. Moreover, the construction of positive and negative pairs in current technologies strongly relies on specific time series characteristics, restricting their generalization across diverse types of time series data. To address these limitations, we propose SimTS, a simple representation learning approach for improving time series forecasting by learning to predict the future from the past in the latent space. SimTS does not rely on negative pairs or specific assumptions about the characteristics of the particular time series. Our extensive experiments on several benchmark time series forecasting datasets show that SimTS achieves competitive performance compared to existing contrastive learning methods. Furthermore, we show the shortcomings of the current contrastive learning framework used for time series forecasting through a detailed ablation study. Overall, our work suggests that SimTS is a promising alternative to other contrastive learning approaches for time series forecasting.
Correlated Product of Experts for Sparse Gaussian Process Regression
Dec 17, 2021
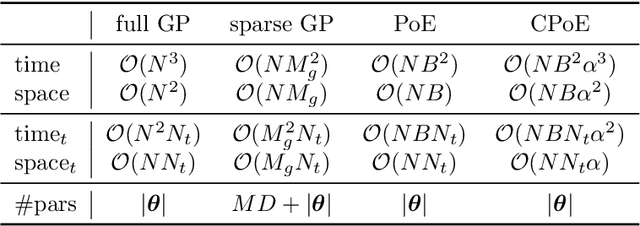

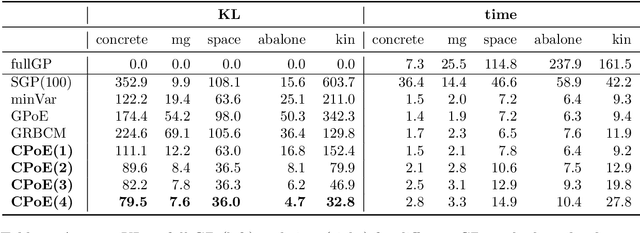
Abstract:Gaussian processes (GPs) are an important tool in machine learning and statistics with applications ranging from social and natural science through engineering. They constitute a powerful kernelized non-parametric method with well-calibrated uncertainty estimates, however, off-the-shelf GP inference procedures are limited to datasets with several thousand data points because of their cubic computational complexity. For this reason, many sparse GPs techniques have been developed over the past years. In this paper, we focus on GP regression tasks and propose a new approach based on aggregating predictions from several local and correlated experts. Thereby, the degree of correlation between the experts can vary between independent up to fully correlated experts. The individual predictions of the experts are aggregated taking into account their correlation resulting in consistent uncertainty estimates. Our method recovers independent Product of Experts, sparse GP and full GP in the limiting cases. The presented framework can deal with a general kernel function and multiple variables, and has a time and space complexity which is linear in the number of experts and data samples, which makes our approach highly scalable. We demonstrate superior performance, in a time vs. accuracy sense, of our proposed method against state-of-the-art GP approximation methods for synthetic as well as several real-world datasets with deterministic and stochastic optimization.
Orthogonally Decoupled Variational Fourier Features
Jul 13, 2020



Abstract:Sparse inducing points have long been a standard method to fit Gaussian processes to big data. In the last few years, spectral methods that exploit approximations of the covariance kernel have shown to be competitive. In this work we exploit a recently introduced orthogonally decoupled variational basis to combine spectral methods and sparse inducing points methods. We show that the method is competitive with the state-of-the-art on synthetic and on real-world data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge