Michael Vollenweider
Fusion of Graph Neural Networks via Optimal Transport
Mar 27, 2025Abstract:In this paper, we explore the idea of combining GCNs into one model. To that end, we align the weights of different models layer-wise using optimal transport (OT). We present and evaluate three types of transportation costs and show that the studied fusion method consistently outperforms the performance of vanilla averaging. Finally, we present results suggesting that model fusion using OT is harder in the case of GCNs than MLPs and that incorporating the graph structure into the process does not improve the performance of the method.
Learning Personalized Treatment Decisions in Precision Medicine: Disentangling Treatment Assignment Bias in Counterfactual Outcome Prediction and Biomarker Identification
Oct 01, 2024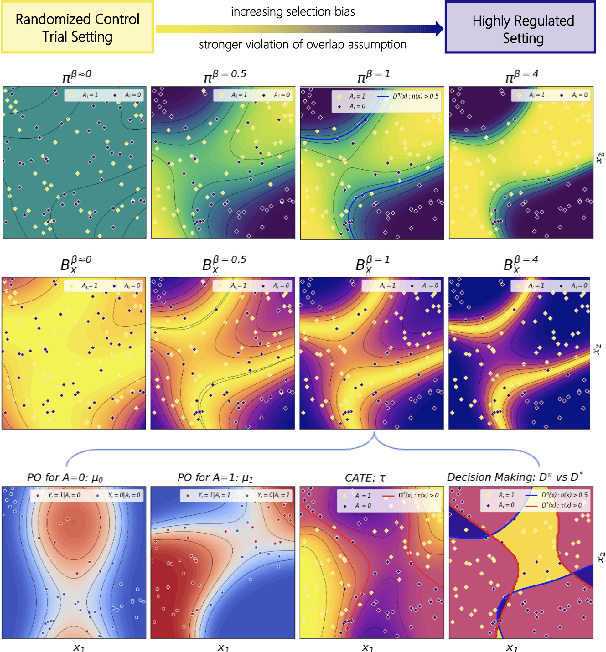
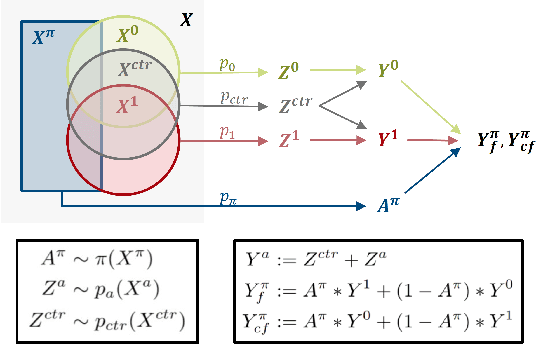
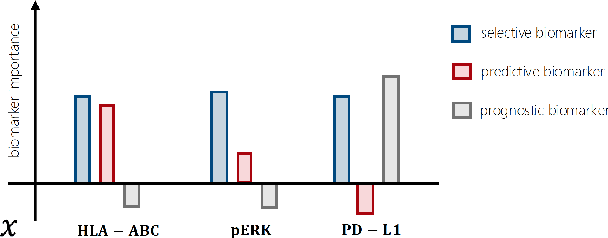
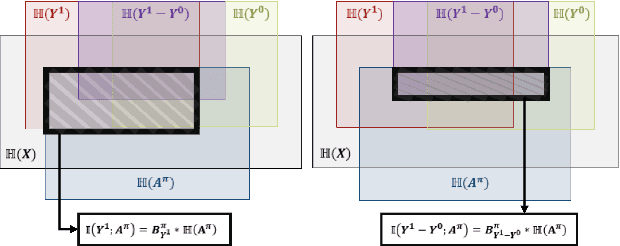
Abstract:Precision medicine offers the potential to tailor treatment decisions to individual patients, yet it faces significant challenges due to the complex biases in clinical observational data and the high-dimensional nature of biological data. This study models various types of treatment assignment biases using mutual information and investigates their impact on machine learning (ML) models for counterfactual prediction and biomarker identification. Unlike traditional counterfactual benchmarks that rely on fixed treatment policies, our work focuses on modeling different characteristics of the underlying observational treatment policy in distinct clinical settings. We validate our approach through experiments on toy datasets, semi-synthetic tumor cancer genome atlas (TCGA) data, and real-world biological outcomes from drug and CRISPR screens. By incorporating empirical biological mechanisms, we create a more realistic benchmark that reflects the complexities of real-world data. Our analysis reveals that different biases lead to varying model performances, with some biases, especially those unrelated to outcome mechanisms, having minimal effect on prediction accuracy. This highlights the crucial need to account for specific biases in clinical observational data in counterfactual ML model development, ultimately enhancing the personalization of treatment decisions in precision medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge