Gabriele Gut
AI-powered virtual tissues from spatial proteomics for clinical diagnostics and biomedical discovery
Jan 10, 2025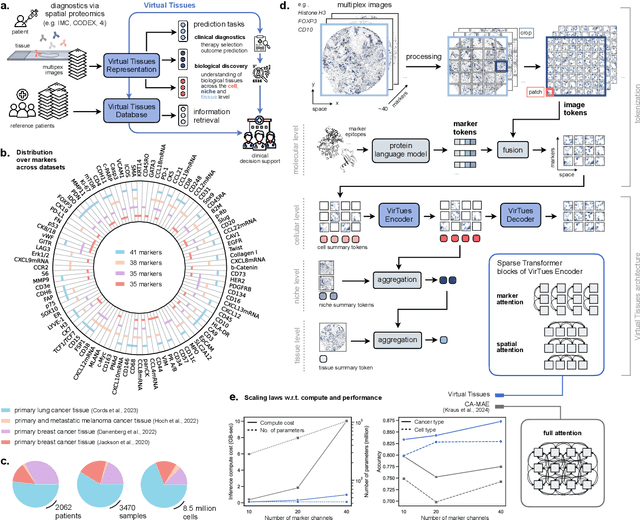
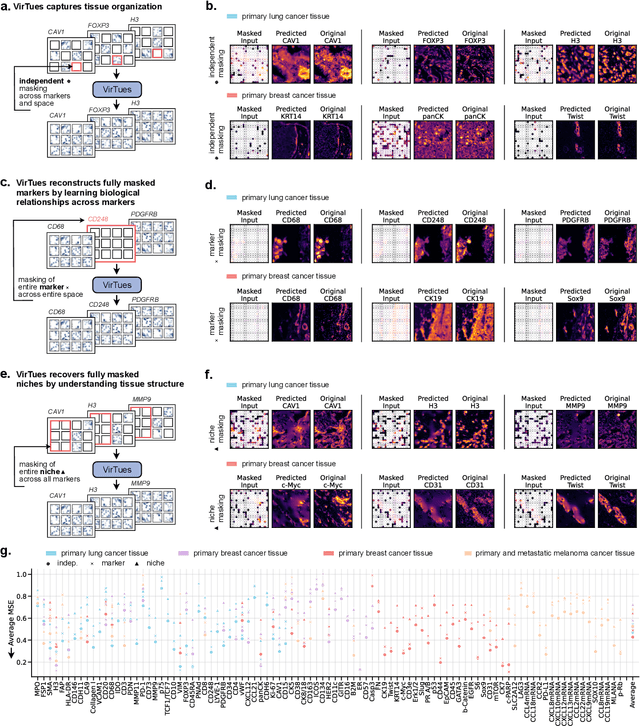
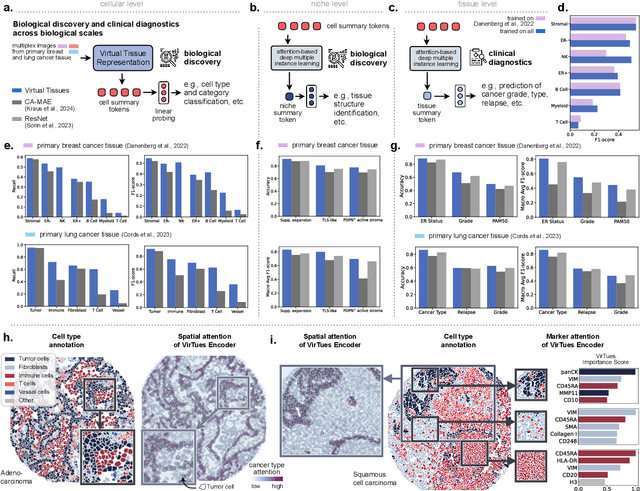
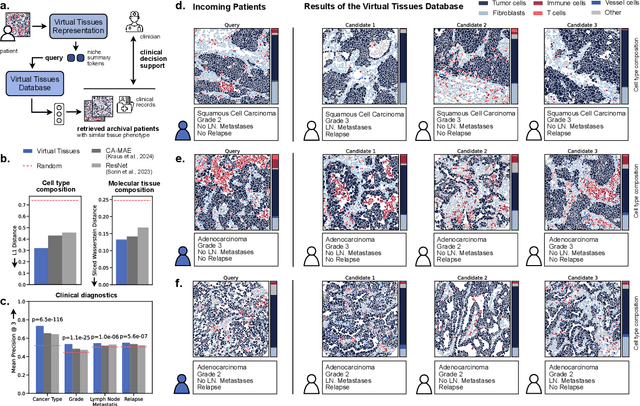
Abstract:Spatial proteomics technologies have transformed our understanding of complex tissue architectures by enabling simultaneous analysis of multiple molecular markers and their spatial organization. The high dimensionality of these data, varying marker combinations across experiments and heterogeneous study designs pose unique challenges for computational analysis. Here, we present Virtual Tissues (VirTues), a foundation model framework for biological tissues that operates across the molecular, cellular and tissue scale. VirTues introduces innovations in transformer architecture design, including a novel tokenization scheme that captures both spatial and marker dimensions, and attention mechanisms that scale to high-dimensional multiplex data while maintaining interpretability. Trained on diverse cancer and non-cancer tissue datasets, VirTues demonstrates strong generalization capabilities without task-specific fine-tuning, enabling cross-study analysis and novel marker integration. As a generalist model, VirTues outperforms existing approaches across clinical diagnostics, biological discovery and patient case retrieval tasks, while providing insights into tissue function and disease mechanisms.
Learning Personalized Treatment Decisions in Precision Medicine: Disentangling Treatment Assignment Bias in Counterfactual Outcome Prediction and Biomarker Identification
Oct 01, 2024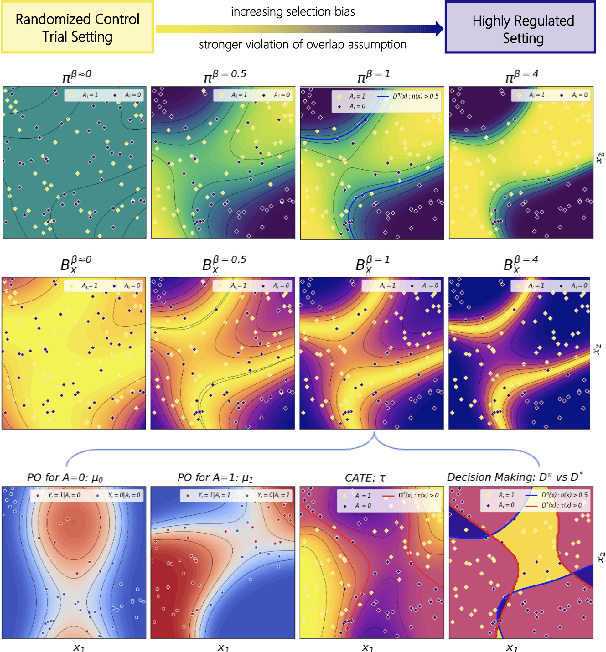
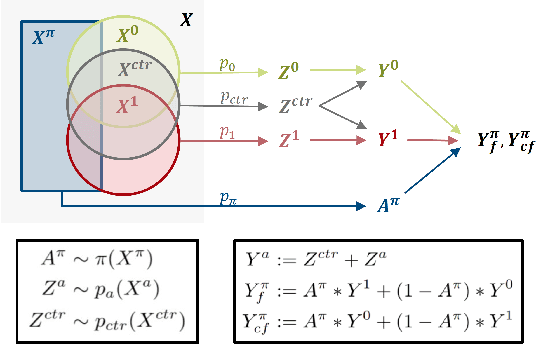
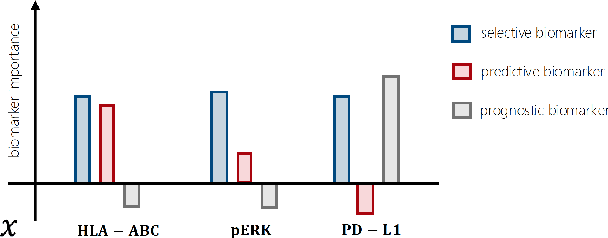
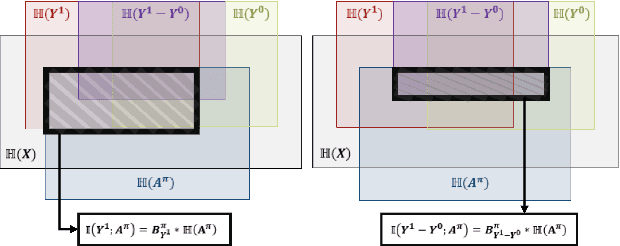
Abstract:Precision medicine offers the potential to tailor treatment decisions to individual patients, yet it faces significant challenges due to the complex biases in clinical observational data and the high-dimensional nature of biological data. This study models various types of treatment assignment biases using mutual information and investigates their impact on machine learning (ML) models for counterfactual prediction and biomarker identification. Unlike traditional counterfactual benchmarks that rely on fixed treatment policies, our work focuses on modeling different characteristics of the underlying observational treatment policy in distinct clinical settings. We validate our approach through experiments on toy datasets, semi-synthetic tumor cancer genome atlas (TCGA) data, and real-world biological outcomes from drug and CRISPR screens. By incorporating empirical biological mechanisms, we create a more realistic benchmark that reflects the complexities of real-world data. Our analysis reveals that different biases lead to varying model performances, with some biases, especially those unrelated to outcome mechanisms, having minimal effect on prediction accuracy. This highlights the crucial need to account for specific biases in clinical observational data in counterfactual ML model development, ultimately enhancing the personalization of treatment decisions in precision medicine.
Towards AI-Based Precision Oncology: A Machine Learning Framework for Personalized Counterfactual Treatment Suggestions based on Multi-Omics Data
Feb 19, 2024



Abstract:AI-driven precision oncology has the transformative potential to reshape cancer treatment by leveraging the power of AI models to analyze the interaction between complex patient characteristics and their corresponding treatment outcomes. New technological platforms have facilitated the timely acquisition of multimodal data on tumor biology at an unprecedented resolution, such as single-cell multi-omics data, making this quality and quantity of data available for data-driven improved clinical decision-making. In this work, we propose a modular machine learning framework designed for personalized counterfactual cancer treatment suggestions based on an ensemble of machine learning experts trained on diverse multi-omics technologies. These specialized counterfactual experts per technology are consistently aggregated into a more powerful expert with superior performance and can provide both confidence and an explanation of its decision. The framework is tailored to address critical challenges inherent in data-driven cancer research, including the high-dimensional nature of the data, and the presence of treatment assignment bias in the retrospective observational data. The framework is showcased through comprehensive demonstrations using data from in-vitro and in-vivo treatment responses from a cohort of patients with ovarian cancer. Our method aims to empower clinicians with a reality-centric decision-support tool including probabilistic treatment suggestions with calibrated confidence and personalized explanations for tailoring treatment strategies to multi-omics characteristics of individual cancer patients.
Neural Unbalanced Optimal Transport via Cycle-Consistent Semi-Couplings
Sep 30, 2022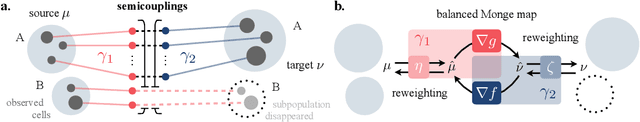
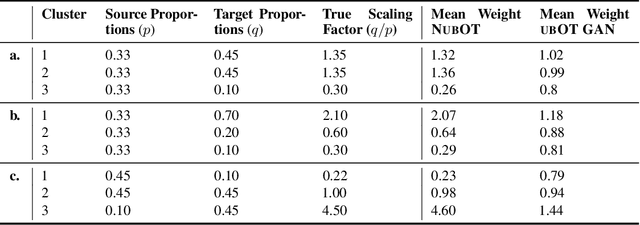

Abstract:Comparing unpaired samples of a distribution or population taken at different points in time is a fundamental task in many application domains where measuring populations is destructive and cannot be done repeatedly on the same sample, such as in single-cell biology. Optimal transport (OT) can solve this challenge by learning an optimal coupling of samples across distributions from unpaired data. However, the usual formulation of OT assumes conservation of mass, which is violated in unbalanced scenarios in which the population size changes (e.g., cell proliferation or death) between measurements. In this work, we introduce NubOT, a neural unbalanced OT formulation that relies on the formalism of semi-couplings to account for creation and destruction of mass. To estimate such semi-couplings and generalize out-of-sample, we derive an efficient parameterization based on neural optimal transport maps and propose a novel algorithmic scheme through a cycle-consistent training procedure. We apply our method to the challenging task of forecasting heterogeneous responses of multiple cancer cell lines to various drugs, where we observe that by accurately modeling cell proliferation and death, our method yields notable improvements over previous neural optimal transport methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge