Johann Wenckstern
AI-powered virtual tissues from spatial proteomics for clinical diagnostics and biomedical discovery
Jan 10, 2025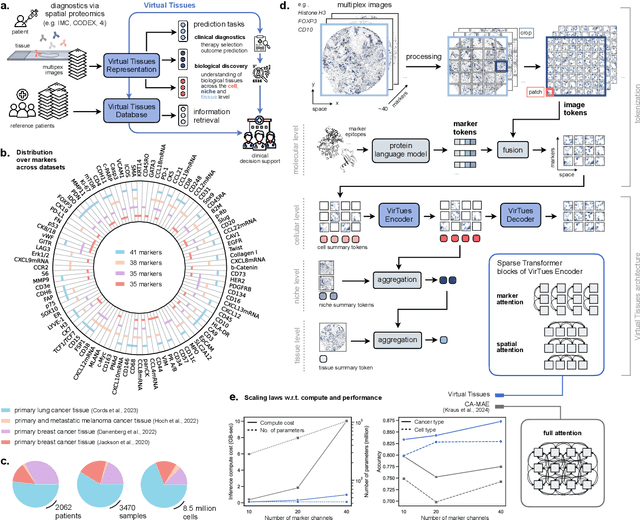
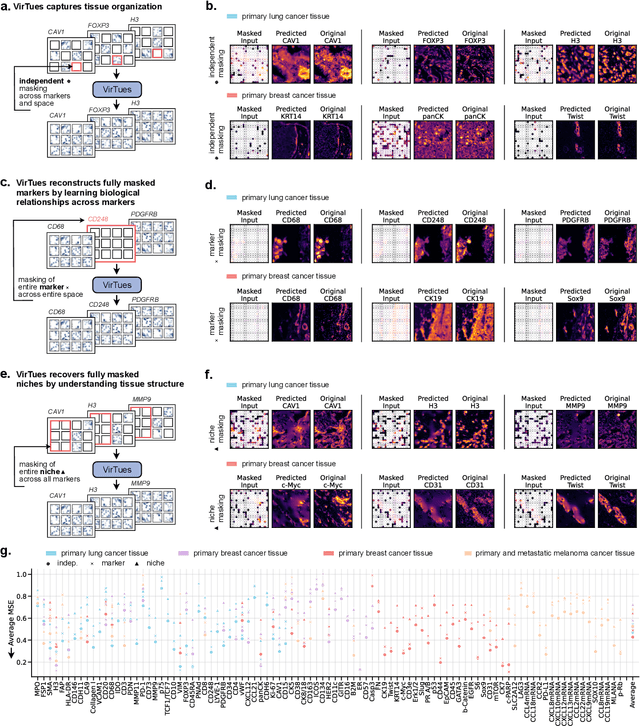
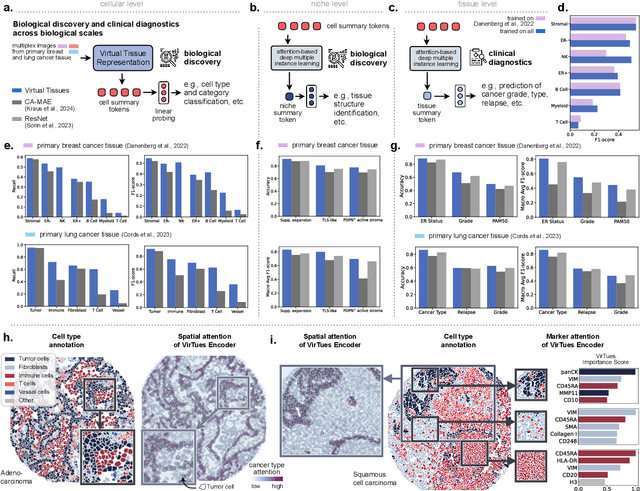
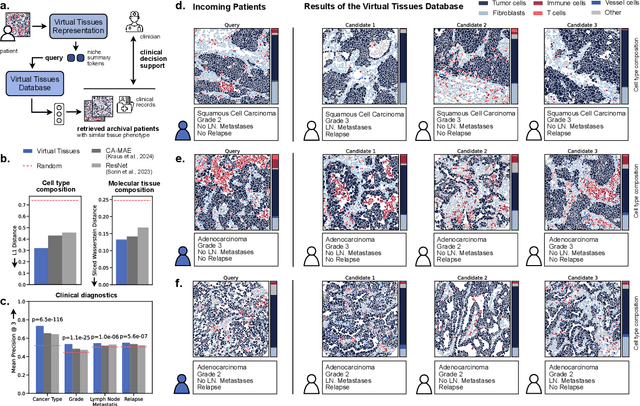
Abstract:Spatial proteomics technologies have transformed our understanding of complex tissue architectures by enabling simultaneous analysis of multiple molecular markers and their spatial organization. The high dimensionality of these data, varying marker combinations across experiments and heterogeneous study designs pose unique challenges for computational analysis. Here, we present Virtual Tissues (VirTues), a foundation model framework for biological tissues that operates across the molecular, cellular and tissue scale. VirTues introduces innovations in transformer architecture design, including a novel tokenization scheme that captures both spatial and marker dimensions, and attention mechanisms that scale to high-dimensional multiplex data while maintaining interpretability. Trained on diverse cancer and non-cancer tissue datasets, VirTues demonstrates strong generalization capabilities without task-specific fine-tuning, enabling cross-study analysis and novel marker integration. As a generalist model, VirTues outperforms existing approaches across clinical diagnostics, biological discovery and patient case retrieval tasks, while providing insights into tissue function and disease mechanisms.
GLAudio Listens to the Sound of the Graph
Jul 19, 2024Abstract:We propose GLAudio: Graph Learning on Audio representation of the node features and the connectivity structure. This novel architecture propagates the node features through the graph network according to the discrete wave equation and then employs a sequence learning architecture to learn the target node function from the audio wave signal. This leads to a new paradigm of learning on graph-structured data, in which information propagation and information processing are separated into two distinct steps. We theoretically characterize the expressivity of our model, introducing the notion of the receptive field of a vertex, and investigate our model's susceptibility to over-smoothing and over-squashing both theoretically as well as experimentally on various graph datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge