Matteo Pariset
AI-powered virtual tissues from spatial proteomics for clinical diagnostics and biomedical discovery
Jan 10, 2025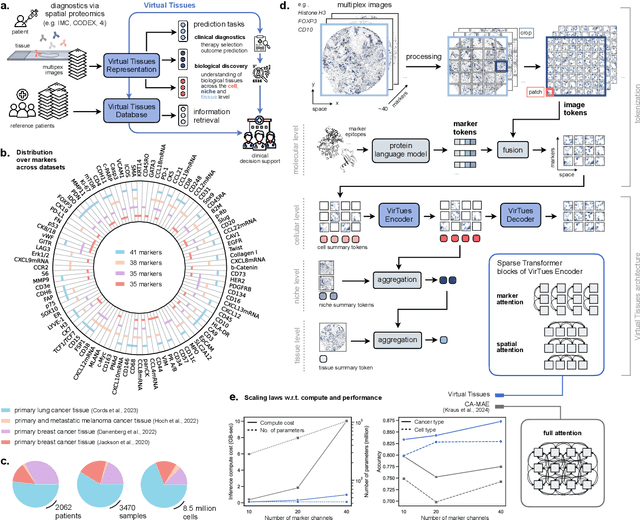
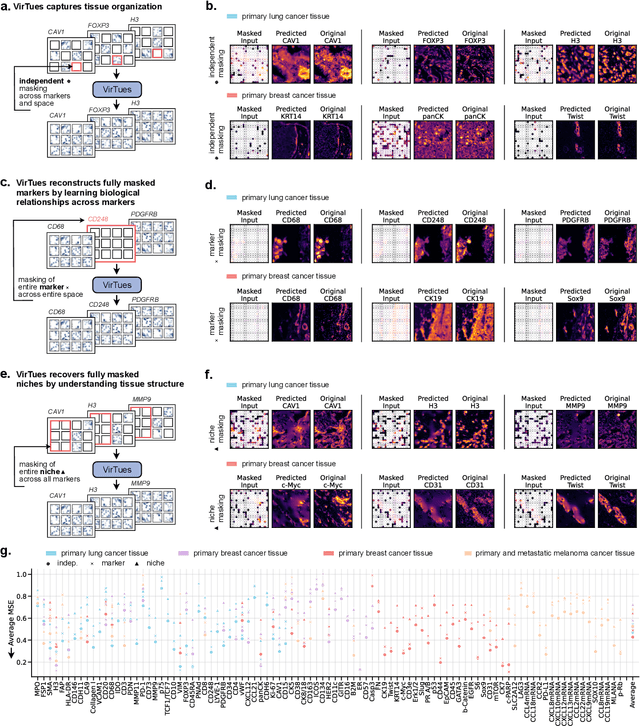
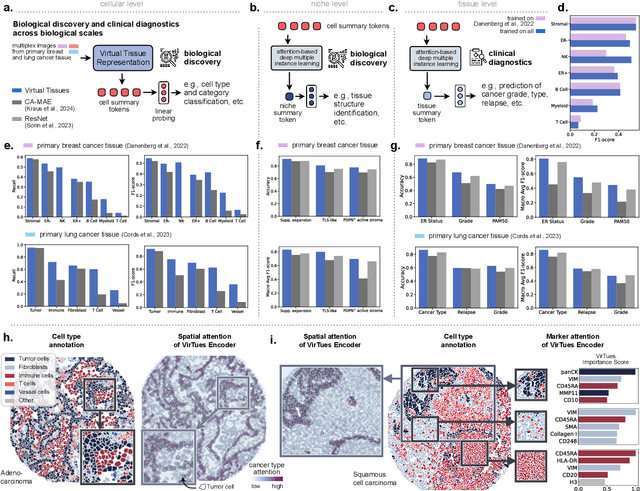
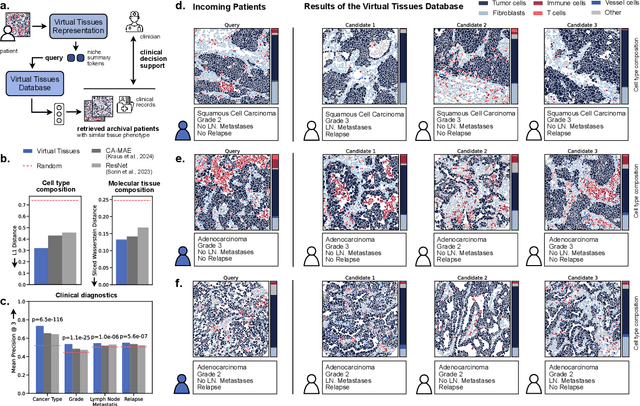
Abstract:Spatial proteomics technologies have transformed our understanding of complex tissue architectures by enabling simultaneous analysis of multiple molecular markers and their spatial organization. The high dimensionality of these data, varying marker combinations across experiments and heterogeneous study designs pose unique challenges for computational analysis. Here, we present Virtual Tissues (VirTues), a foundation model framework for biological tissues that operates across the molecular, cellular and tissue scale. VirTues introduces innovations in transformer architecture design, including a novel tokenization scheme that captures both spatial and marker dimensions, and attention mechanisms that scale to high-dimensional multiplex data while maintaining interpretability. Trained on diverse cancer and non-cancer tissue datasets, VirTues demonstrates strong generalization capabilities without task-specific fine-tuning, enabling cross-study analysis and novel marker integration. As a generalist model, VirTues outperforms existing approaches across clinical diagnostics, biological discovery and patient case retrieval tasks, while providing insights into tissue function and disease mechanisms.
Unbalanced Diffusion Schrödinger Bridge
Jun 15, 2023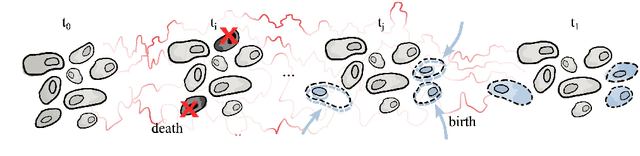

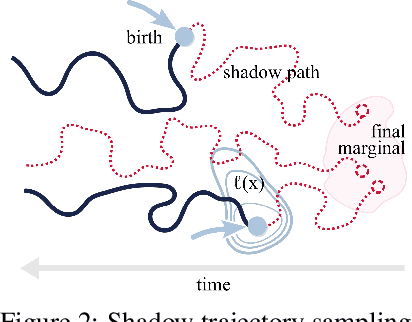

Abstract:Schr\"odinger bridges (SBs) provide an elegant framework for modeling the temporal evolution of populations in physical, chemical, or biological systems. Such natural processes are commonly subject to changes in population size over time due to the emergence of new species or birth and death events. However, existing neural parameterizations of SBs such as diffusion Schr\"odinger bridges (DSBs) are restricted to settings in which the endpoints of the stochastic process are both probability measures and assume conservation of mass constraints. To address this limitation, we introduce unbalanced DSBs which model the temporal evolution of marginals with arbitrary finite mass. This is achieved by deriving the time reversal of stochastic differential equations with killing and birth terms. We present two novel algorithmic schemes that comprise a scalable objective function for training unbalanced DSBs and provide a theoretical analysis alongside challenging applications on predicting heterogeneous molecular single-cell responses to various cancer drugs and simulating the emergence and spread of new viral variants.
Aligned Diffusion Schrödinger Bridges
Feb 22, 2023Abstract:Diffusion Schr\"odinger bridges (DSB) have recently emerged as a powerful framework for recovering stochastic dynamics via their marginal observations at different time points. Despite numerous successful applications, existing algorithms for solving DSBs have so far failed to utilize the structure of aligned data, which naturally arises in many biological phenomena. In this paper, we propose a novel algorithmic framework that, for the first time, solves DSBs while respecting the data alignment. Our approach hinges on a combination of two decades-old ideas: The classical Schr\"odinger bridge theory and Doob's $h$-transform. Compared to prior methods, our approach leads to a simpler training procedure with lower variance, which we further augment with principled regularization schemes. This ultimately leads to sizeable improvements across experiments on synthetic and real data, including the tasks of rigid protein docking and temporal evolution of cellular differentiation processes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge