Eeshaan Jain
AI-powered virtual tissues from spatial proteomics for clinical diagnostics and biomedical discovery
Jan 10, 2025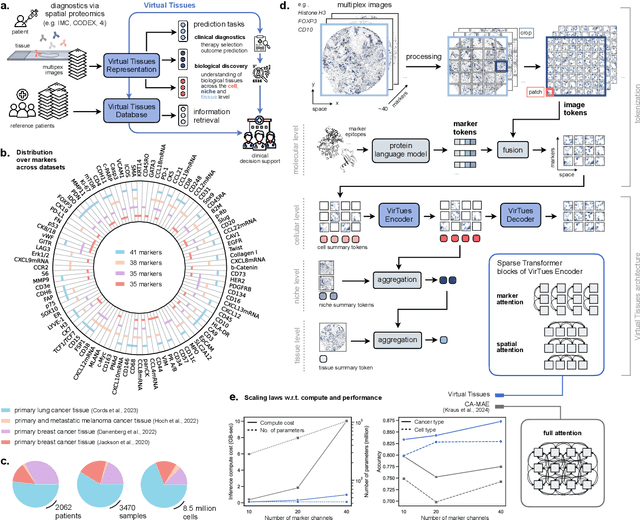
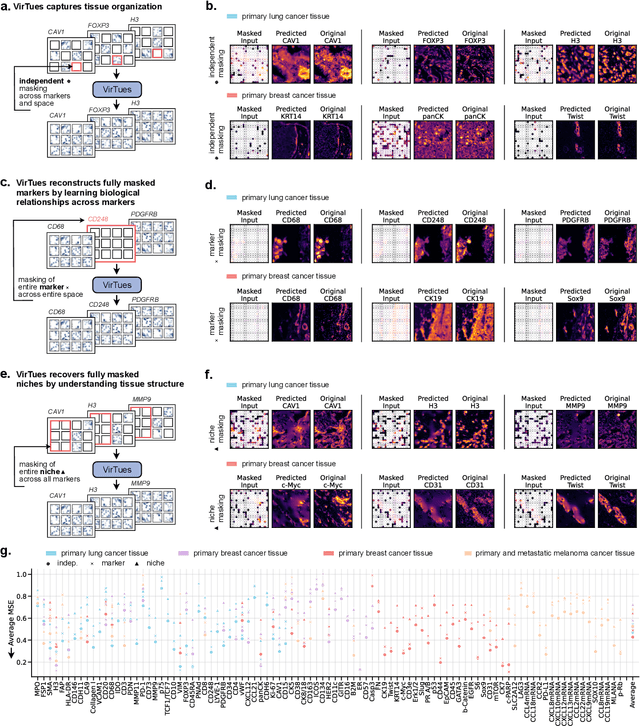
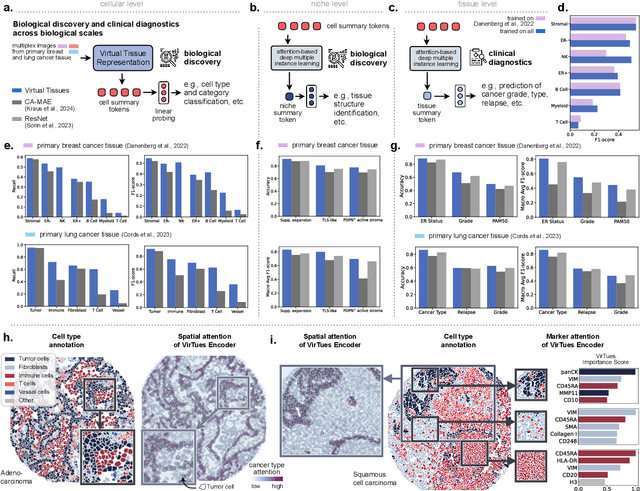
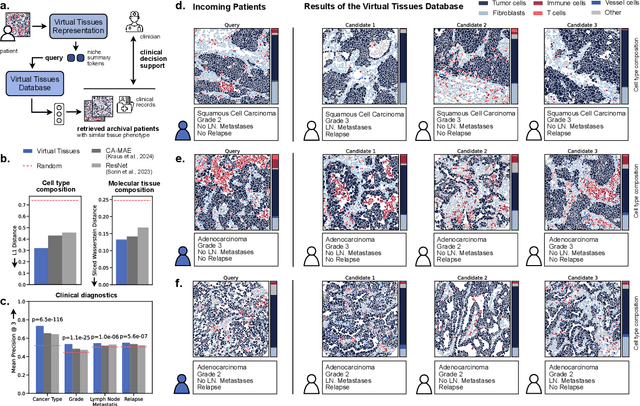
Abstract:Spatial proteomics technologies have transformed our understanding of complex tissue architectures by enabling simultaneous analysis of multiple molecular markers and their spatial organization. The high dimensionality of these data, varying marker combinations across experiments and heterogeneous study designs pose unique challenges for computational analysis. Here, we present Virtual Tissues (VirTues), a foundation model framework for biological tissues that operates across the molecular, cellular and tissue scale. VirTues introduces innovations in transformer architecture design, including a novel tokenization scheme that captures both spatial and marker dimensions, and attention mechanisms that scale to high-dimensional multiplex data while maintaining interpretability. Trained on diverse cancer and non-cancer tissue datasets, VirTues demonstrates strong generalization capabilities without task-specific fine-tuning, enabling cross-study analysis and novel marker integration. As a generalist model, VirTues outperforms existing approaches across clinical diagnostics, biological discovery and patient case retrieval tasks, while providing insights into tissue function and disease mechanisms.
Graph Edit Distance with General Costs Using Neural Set Divergence
Sep 26, 2024



Abstract:Graph Edit Distance (GED) measures the (dis-)similarity between two given graphs, in terms of the minimum-cost edit sequence that transforms one graph to the other. However, the exact computation of GED is NP-Hard, which has recently motivated the design of neural methods for GED estimation. However, they do not explicitly account for edit operations with different costs. In response, we propose GRAPHEDX, a neural GED estimator that can work with general costs specified for the four edit operations, viz., edge deletion, edge addition, node deletion and node addition. We first present GED as a quadratic assignment problem (QAP) that incorporates these four costs. Then, we represent each graph as a set of node and edge embeddings and use them to design a family of neural set divergence surrogates. We replace the QAP terms corresponding to each operation with their surrogates. Computing such neural set divergence require aligning nodes and edges of the two graphs. We learn these alignments using a Gumbel-Sinkhorn permutation generator, additionally ensuring that the node and edge alignments are consistent with each other. Moreover, these alignments are cognizant of both the presence and absence of edges between node-pairs. Experiments on several datasets, under a variety of edit cost settings, show that GRAPHEDX consistently outperforms state-of-the-art methods and heuristics in terms of prediction error.
* Published at NeurIPS 2024
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge