Stefano Mensa
Qiskit Machine Learning: an open-source library for quantum machine learning tasks at scale on quantum hardware and classical simulators
May 23, 2025Abstract:We present Qiskit Machine Learning (ML), a high-level Python library that combines elements of quantum computing with traditional machine learning. The API abstracts Qiskit's primitives to facilitate interactions with classical simulators and quantum hardware. Qiskit ML started as a proof-of-concept code in 2019 and has since been developed to be a modular, intuitive tool for non-specialist users while allowing extensibility and fine-tuning controls for quantum computational scientists and developers. The library is available as a public, open-source tool and is distributed under the Apache version 2.0 license.
Evolving a Multi-Population Evolutionary-QAOA on Distributed QPUs
Sep 19, 2024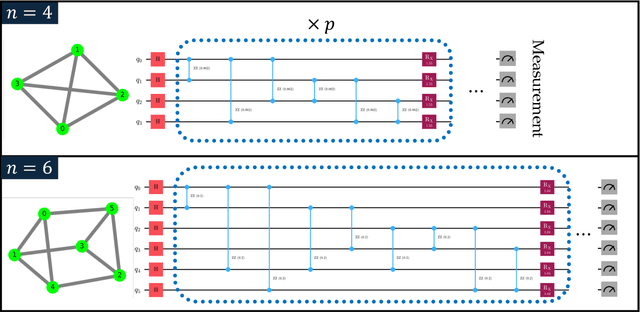
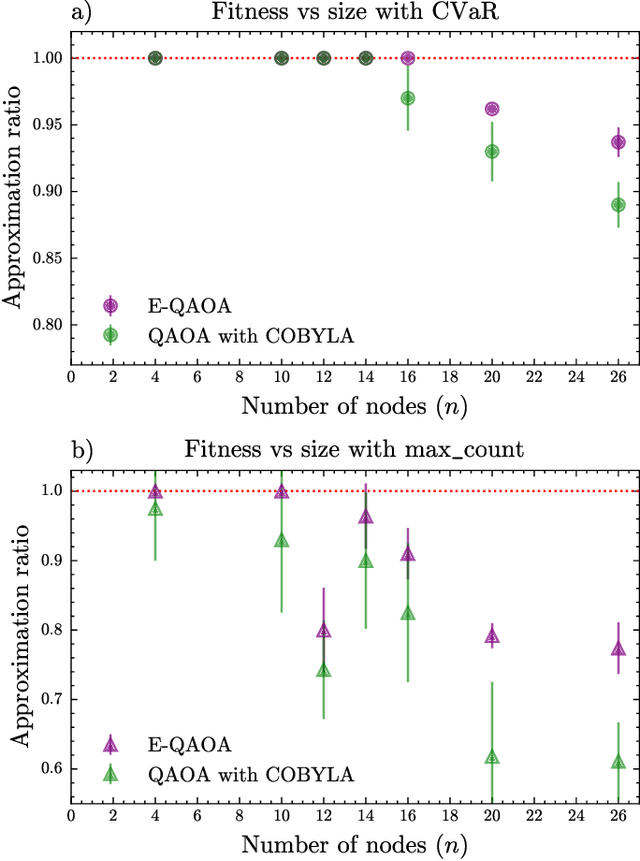
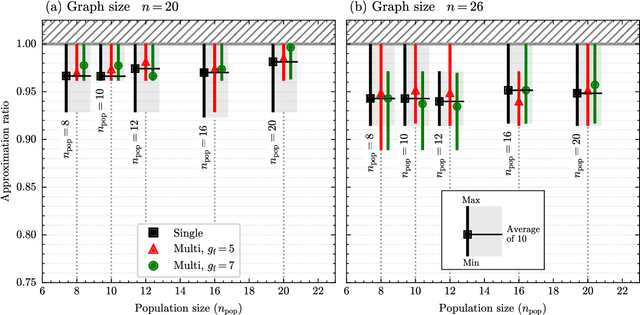
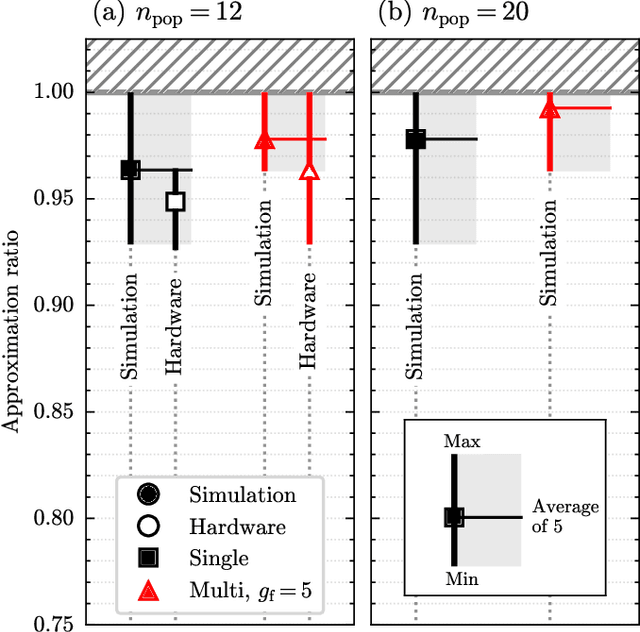
Abstract:Our research combines an Evolutionary Algorithm (EA) with a Quantum Approximate Optimization Algorithm (QAOA) to update the ansatz parameters, in place of traditional gradient-based methods, and benchmark on the Max-Cut problem. We demonstrate that our Evolutionary-QAOA (E-QAOA) pairing performs on par or better than a COBYLA-based QAOA in terms of solution accuracy and variance, for $d$-3 regular graphs between 4 and 26 nodes, using both $max\_count$ and Conditional Value at Risk (CVaR) for fitness function evaluations. Furthermore, we take our algorithm one step further and present a novel approach by presenting a multi-population EA distributed on two QPUs, which evolves independent and isolated populations in parallel, classically communicating elite individuals. Experiments were conducted on both simulators and IBM quantum hardware, and we investigated the relative performance accuracy and variance.
Efficient Parameter Optimisation for Quantum Kernel Alignment: A Sub-sampling Approach in Variational Training
Jan 05, 2024



Abstract:Quantum machine learning with quantum kernels for classification problems is a growing area of research. Recently, quantum kernel alignment techniques that parameterise the kernel have been developed, allowing the kernel to be trained and therefore aligned with a specific dataset. While quantum kernel alignment is a promising technique, it has been hampered by considerable training costs because the full kernel matrix must be constructed at every training iteration. Addressing this challenge, we introduce a novel method that seeks to balance efficiency and performance. We present a sub-sampling training approach that uses a subset of the kernel matrix at each training step, thereby reducing the overall computational cost of the training. In this work, we apply the sub-sampling method to synthetic datasets and a real-world breast cancer dataset and demonstrate considerable reductions in the number of circuits required to train the quantum kernel while maintaining classification accuracy.
Quantum Machine Learning Framework for Virtual Screening in Drug Discovery: a Prospective Quantum Advantage
Apr 08, 2022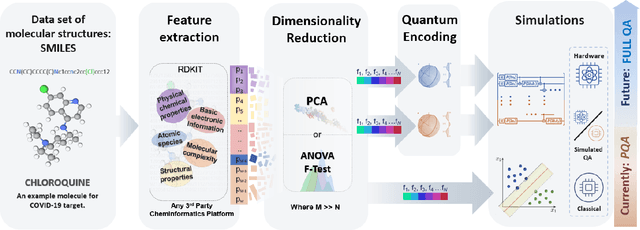
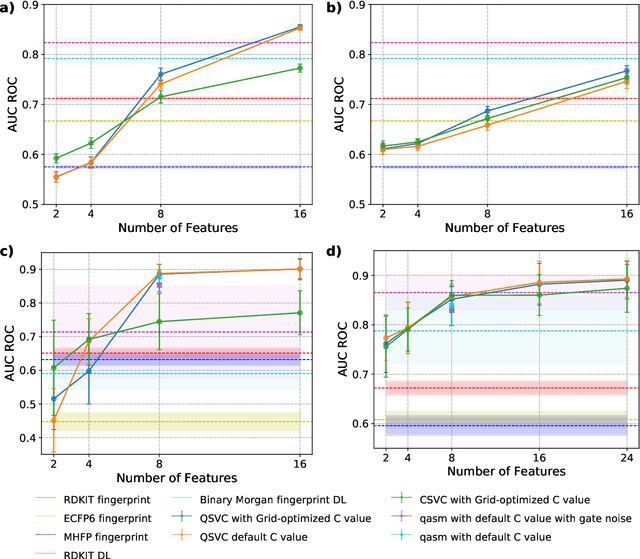
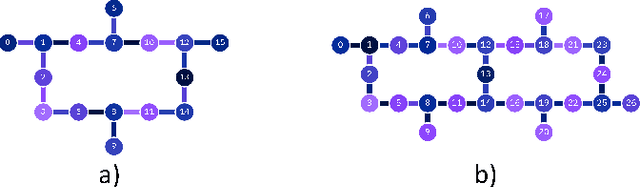
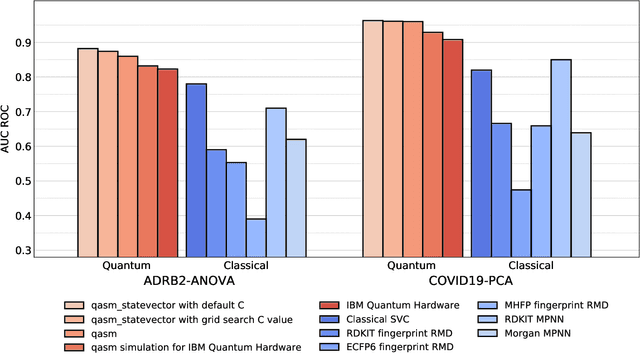
Abstract:Machine Learning (ML) for Ligand Based Virtual Screening (LB-VS) is an important in-silico tool for discovering new drugs in a faster and cost-effective manner, especially for emerging diseases such as COVID-19. In this paper, we propose a general-purpose framework combining a classical Support Vector Classifier (SVC) algorithm with quantum kernel estimation for LB-VS on real-world databases, and we argue in favor of its prospective quantum advantage. Indeed, we heuristically prove that our quantum integrated workflow can, at least in some relevant instances, provide a tangible advantage compared to state-of-art classical algorithms operating on the same datasets, showing strong dependence on target and features selection method. Finally, we test our algorithm on IBM Quantum processors using ADRB2 and COVID-19 datasets, showing that hardware simulations provide results in line with the predicted performances and can surpass classical equivalents.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge