Kevin Thandiackal
IBM Research Europe, ETH Zurich
Multi-scale Feature Alignment for Continual Learning of Unlabeled Domains
Feb 02, 2023Abstract:Methods for unsupervised domain adaptation (UDA) help to improve the performance of deep neural networks on unseen domains without any labeled data. Especially in medical disciplines such as histopathology, this is crucial since large datasets with detailed annotations are scarce. While the majority of existing UDA methods focus on the adaptation from a labeled source to a single unlabeled target domain, many real-world applications with a long life cycle involve more than one target domain. Thus, the ability to sequentially adapt to multiple target domains becomes essential. In settings where the data from previously seen domains cannot be stored, e.g., due to data protection regulations, the above becomes a challenging continual learning problem. To this end, we propose to use generative feature-driven image replay in conjunction with a dual-purpose discriminator that not only enables the generation of images with realistic features for replay, but also promotes feature alignment during domain adaptation. We evaluate our approach extensively on a sequence of three histopathological datasets for tissue-type classification, achieving state-of-the-art results. We present detailed ablation experiments studying our proposed method components and demonstrate a possible use-case of our continual UDA method for an unsupervised patch-based segmentation task given high-resolution tissue images.
Weakly Supervised Joint Whole-Slide Segmentation and Classification in Prostate Cancer
Jan 07, 2023Abstract:The segmentation and automatic identification of histological regions of diagnostic interest offer a valuable aid to pathologists. However, segmentation methods are hampered by the difficulty of obtaining pixel-level annotations, which are tedious and expensive to obtain for Whole-Slide images (WSI). To remedy this, weakly supervised methods have been developed to exploit the annotations directly available at the image level. However, to our knowledge, none of these techniques is adapted to deal with WSIs. In this paper, we propose WholeSIGHT, a weakly-supervised method, to simultaneously segment and classify WSIs of arbitrary shapes and sizes. Formally, WholeSIGHT first constructs a tissue-graph representation of the WSI, where the nodes and edges depict tissue regions and their interactions, respectively. During training, a graph classification head classifies the WSI and produces node-level pseudo labels via post-hoc feature attribution. These pseudo labels are then used to train a node classification head for WSI segmentation. During testing, both heads simultaneously render class prediction and segmentation for an input WSI. We evaluated WholeSIGHT on three public prostate cancer WSI datasets. Our method achieved state-of-the-art weakly-supervised segmentation performance on all datasets while resulting in better or comparable classification with respect to state-of-the-art weakly-supervised WSI classification methods. Additionally, we quantify the generalization capability of our method in terms of segmentation and classification performance, uncertainty estimation, and model calibration.
Generative appearance replay for continual unsupervised domain adaptation
Jan 03, 2023



Abstract:Deep learning models can achieve high accuracy when trained on large amounts of labeled data. However, real-world scenarios often involve several challenges: Training data may become available in installments, may originate from multiple different domains, and may not contain labels for training. Certain settings, for instance medical applications, often involve further restrictions that prohibit retention of previously seen data due to privacy regulations. In this work, to address such challenges, we study unsupervised segmentation in continual learning scenarios that involve domain shift. To that end, we introduce GarDA (Generative Appearance Replay for continual Domain Adaptation), a generative-replay based approach that can adapt a segmentation model sequentially to new domains with unlabeled data. In contrast to single-step unsupervised domain adaptation (UDA), continual adaptation to a sequence of domains enables leveraging and consolidation of information from multiple domains. Unlike previous approaches in incremental UDA, our method does not require access to previously seen data, making it applicable in many practical scenarios. We evaluate GarDA on two datasets with different organs and modalities, where it substantially outperforms existing techniques.
Differentiable Zooming for Multiple Instance Learning on Whole-Slide Images
Apr 30, 2022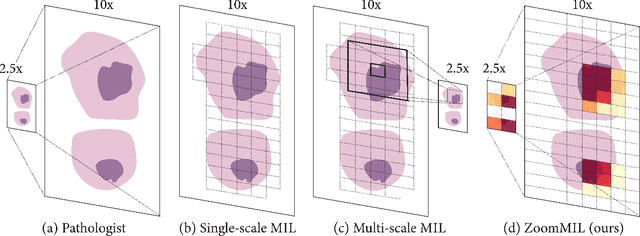
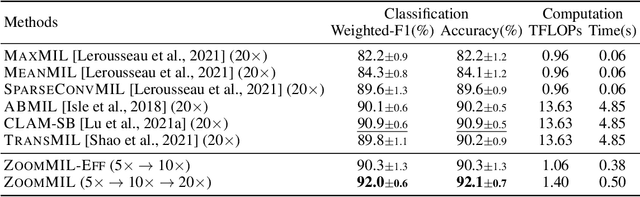
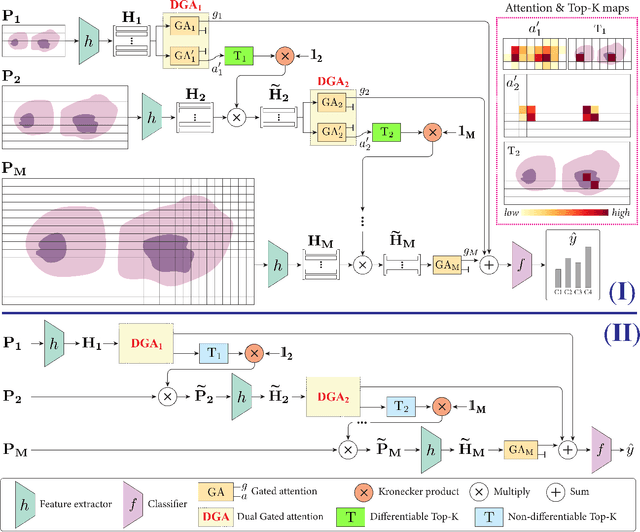
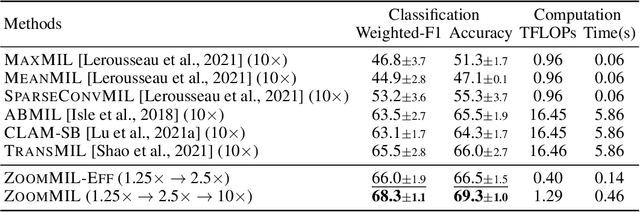
Abstract:Multiple Instance Learning (MIL) methods have become increasingly popular for classifying giga-pixel sized Whole-Slide Images (WSIs) in digital pathology. Most MIL methods operate at a single WSI magnification, by processing all the tissue patches. Such a formulation induces high computational requirements, and constrains the contextualization of the WSI-level representation to a single scale. A few MIL methods extend to multiple scales, but are computationally more demanding. In this paper, inspired by the pathological diagnostic process, we propose ZoomMIL, a method that learns to perform multi-level zooming in an end-to-end manner. ZoomMIL builds WSI representations by aggregating tissue-context information from multiple magnifications. The proposed method outperforms the state-of-the-art MIL methods in WSI classification on two large datasets, while significantly reducing the computational demands with regard to Floating-Point Operations (FLOPs) and processing time by up to 40x.
Match What Matters: Generative Implicit Feature Replay for Continual Learning
Jun 09, 2021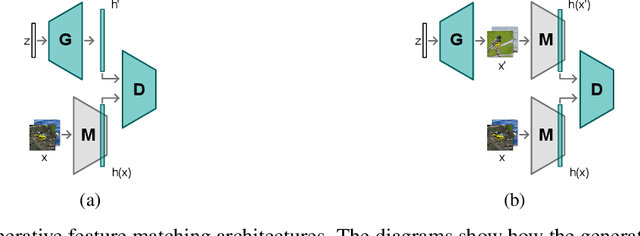
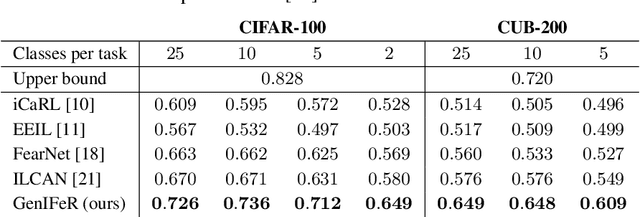
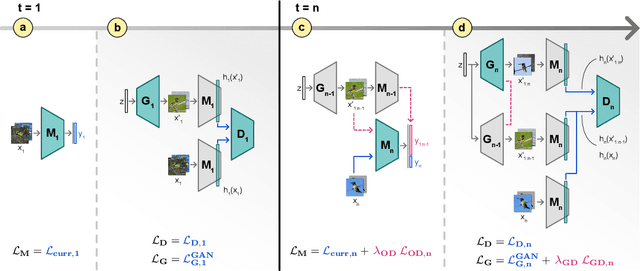

Abstract:Neural networks are prone to catastrophic forgetting when trained incrementally on different tasks. In order to prevent forgetting, most existing methods retain a small subset of previously seen samples, which in turn can be used for joint training with new tasks. While this is indeed effective, it may not always be possible to store such samples, e.g., due to data protection regulations. In these cases, one can instead employ generative models to create artificial samples or features representing memories from previous tasks. Following a similar direction, we propose GenIFeR (Generative Implicit Feature Replay) for class-incremental learning. The main idea is to train a generative adversarial network (GAN) to generate images that contain realistic features. While the generator creates images at full resolution, the discriminator only sees the corresponding features extracted by the continually trained classifier. Since the classifier compresses raw images into features that are actually relevant for classification, the GAN can match this target distribution more accurately. On the other hand, allowing the generator to create full resolution images has several benefits: In contrast to previous approaches, the feature extractor of the classifier does not have to be frozen. In addition, we can employ augmentations on generated images, which not only boosts classification performance, but also mitigates discriminator overfitting during GAN training. We empirically show that GenIFeR is superior to both conventional generative image and feature replay. In particular, we significantly outperform the state-of-the-art in generative replay for various settings on the CIFAR-100 and CUB-200 datasets.
Artificial Intelligence Decision Support for Medical Triage
Nov 09, 2020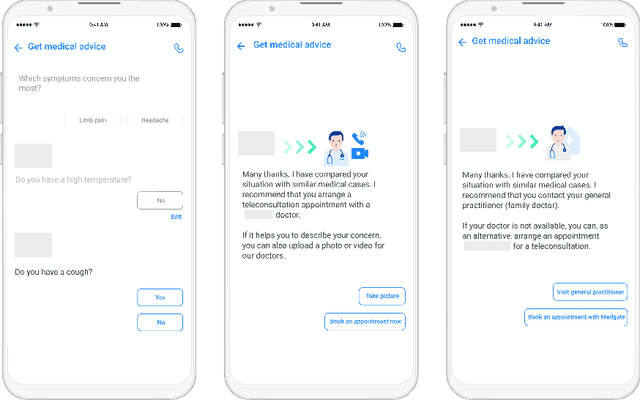
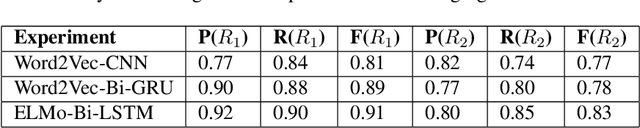
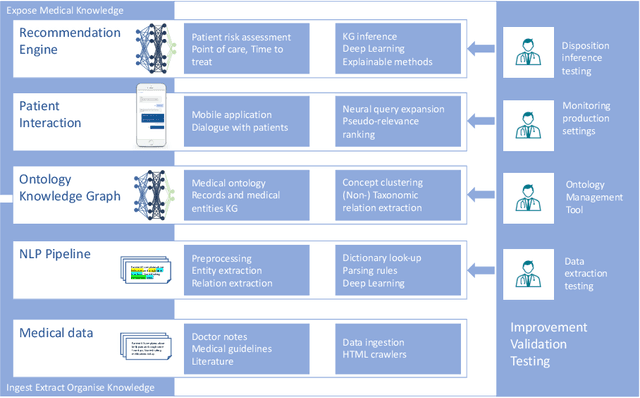
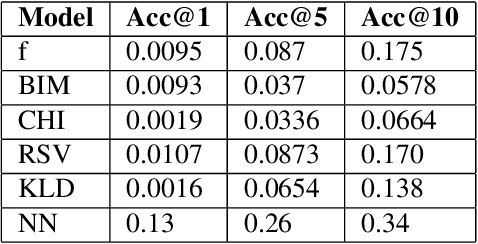
Abstract:Applying state-of-the-art machine learning and natural language processing on approximately one million of teleconsultation records, we developed a triage system, now certified and in use at the largest European telemedicine provider. The system evaluates care alternatives through interactions with patients via a mobile application. Reasoning on an initial set of provided symptoms, the triage application generates AI-powered, personalized questions to better characterize the problem and recommends the most appropriate point of care and time frame for a consultation. The underlying technology was developed to meet the needs for performance, transparency, user acceptance and ease of use, central aspects to the adoption of AI-based decision support systems. Providing such remote guidance at the beginning of the chain of care has significant potential for improving cost efficiency, patient experience and outcomes. Being remote, always available and highly scalable, this service is fundamental in high demand situations, such as the current COVID-19 outbreak.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge