Tiziano Portenier
ETH Zurich
Unpaired Translation from Semantic Label Maps to Images by Leveraging Domain-Specific Simulations
Feb 21, 2023Abstract:Photorealistic image generation from simulated label maps are necessitated in several contexts, such as for medical training in virtual reality. With conventional deep learning methods, this task requires images that are paired with semantic annotations, which typically are unavailable. We introduce a contrastive learning framework for generating photorealistic images from simulated label maps, by learning from unpaired sets of both. Due to potentially large scene differences between real images and label maps, existing unpaired image translation methods lead to artifacts of scene modification in synthesized images. We utilize simulated images as surrogate targets for a contrastive loss, while ensuring consistency by utilizing features from a reverse translation network. Our method enables bidirectional label-image translations, which is demonstrated in a variety of scenarios and datasets, including laparoscopy, ultrasound, and driving scenes. By comparing with state-of-the-art unpaired translation methods, our proposed method is shown to generate realistic and scene-accurate translations.
Match What Matters: Generative Implicit Feature Replay for Continual Learning
Jun 09, 2021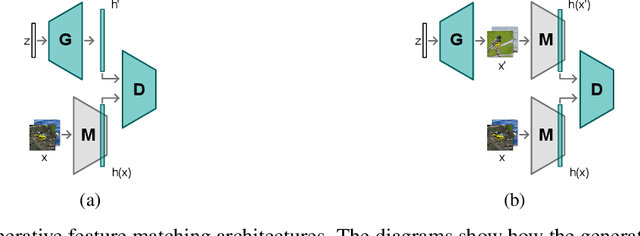
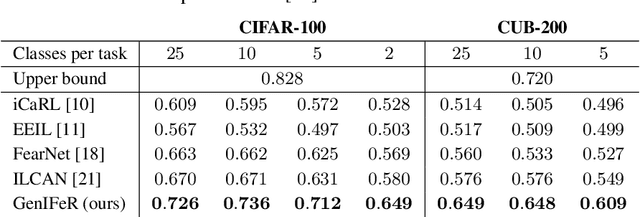
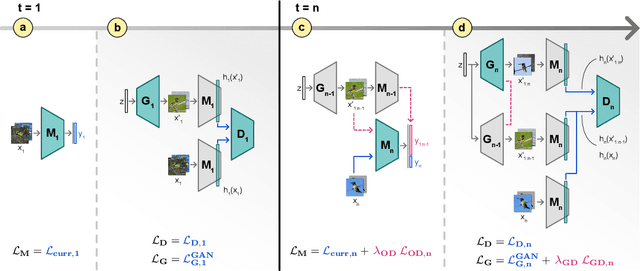

Abstract:Neural networks are prone to catastrophic forgetting when trained incrementally on different tasks. In order to prevent forgetting, most existing methods retain a small subset of previously seen samples, which in turn can be used for joint training with new tasks. While this is indeed effective, it may not always be possible to store such samples, e.g., due to data protection regulations. In these cases, one can instead employ generative models to create artificial samples or features representing memories from previous tasks. Following a similar direction, we propose GenIFeR (Generative Implicit Feature Replay) for class-incremental learning. The main idea is to train a generative adversarial network (GAN) to generate images that contain realistic features. While the generator creates images at full resolution, the discriminator only sees the corresponding features extracted by the continually trained classifier. Since the classifier compresses raw images into features that are actually relevant for classification, the GAN can match this target distribution more accurately. On the other hand, allowing the generator to create full resolution images has several benefits: In contrast to previous approaches, the feature extractor of the classifier does not have to be frozen. In addition, we can employ augmentations on generated images, which not only boosts classification performance, but also mitigates discriminator overfitting during GAN training. We empirically show that GenIFeR is superior to both conventional generative image and feature replay. In particular, we significantly outperform the state-of-the-art in generative replay for various settings on the CIFAR-100 and CUB-200 datasets.
Content-Preserving Unpaired Translation from Simulated to Realistic Ultrasound Images
Mar 09, 2021



Abstract:Interactive simulation of ultrasound imaging greatly facilitates sonography training. Although ray-tracing based methods have shown promising results, obtaining realistic images requires substantial modeling effort and manual parameter tuning. In addition, current techniques still result in a significant appearance gap between simulated images and real clinical scans. In this work we introduce a novel image translation framework to bridge this appearance gap, while preserving the anatomical layout of the simulated scenes. We achieve this goal by leveraging both simulated images with semantic segmentations and unpaired in-vivo ultrasound scans. Our framework is based on recent contrastive unpaired translation techniques and we propose a regularization approach by learning an auxiliary segmentation-to-real image translation task, which encourages the disentanglement of content and style. In addition, we extend the generator to be class-conditional, which enables the incorporation of additional losses, in particular a cyclic consistency loss, to further improve the translation quality. Qualitative and quantitative comparisons against state-of-the-art unpaired translation methods demonstrate the superiority of our proposed framework.
Probabilistic Spatial Analysis in Quantitative Microscopy with Uncertainty-Aware Cell Detection using Deep Bayesian Regression of Density Maps
Feb 23, 2021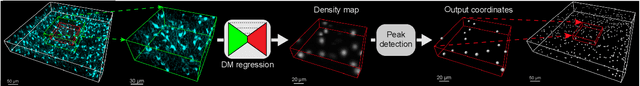

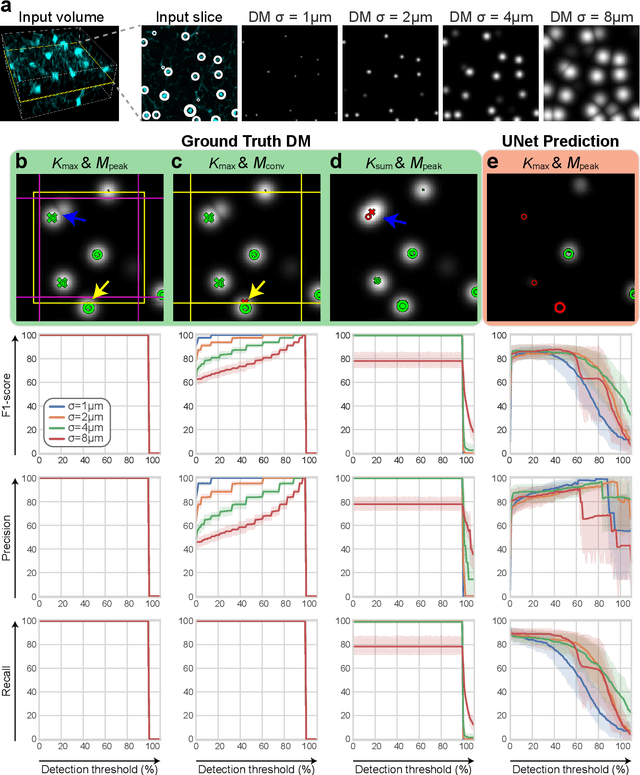
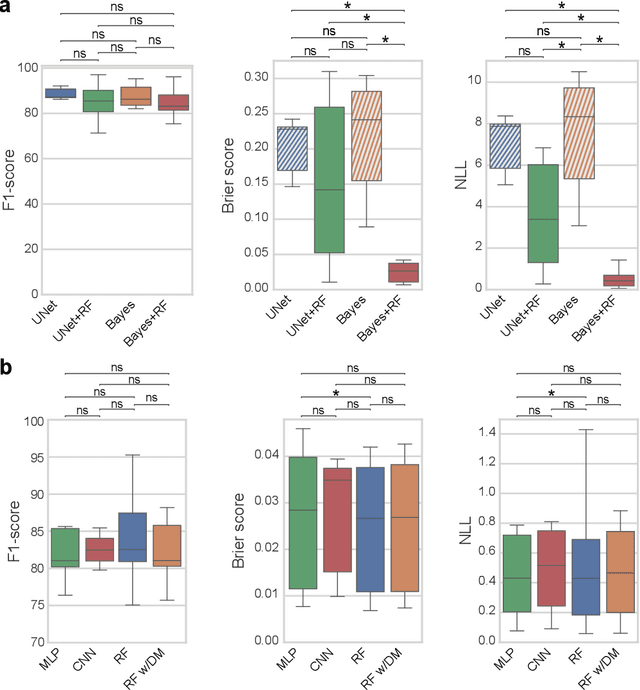
Abstract:3D microscopy is key in the investigation of diverse biological systems, and the ever increasing availability of large datasets demands automatic cell identification methods that not only are accurate, but also can imply the uncertainty in their predictions to inform about potential errors and hence confidence in conclusions using them. While conventional deep learning methods often yield deterministic results, advances in deep Bayesian learning allow for accurate predictions with a probabilistic interpretation in numerous image classification and segmentation tasks. It is however nontrivial to extend such Bayesian methods to cell detection, which requires specialized learning frameworks. In particular, regression of density maps is a popular successful approach for extracting cell coordinates from local peaks in a postprocessing step, which hinders any meaningful probabilistic output. We herein propose a deep learning-based cell detection framework that can operate on large microscopy images and outputs desired probabilistic predictions by (i) integrating Bayesian techniques for the regression of uncertainty-aware density maps, where peak detection can be applied to generate cell proposals, and (ii) learning a mapping from the numerous proposals to a probabilistic space that is calibrated, i.e. accurately represents the chances of a successful prediction. Utilizing such calibrated predictions, we propose a probabilistic spatial analysis with Monte-Carlo sampling. We demonstrate this in revising an existing description of the distribution of a mesenchymal stromal cell type within the bone marrow, where our proposed methods allow us to reveal spatial patterns that are otherwise undetectable. Introducing such probabilistic analysis in quantitative microscopy pipelines will allow for reporting confidence intervals for testing biological hypotheses of spatial distributions.
Utilizing Uncertainty Estimation in Deep Learning Segmentation of Fluorescence Microscopy Images with Missing Markers
Jan 27, 2021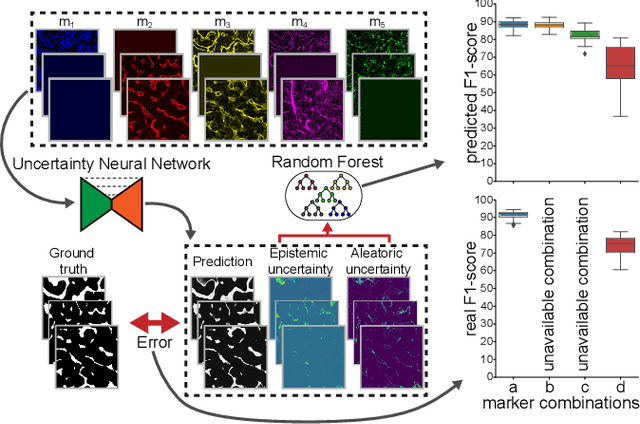
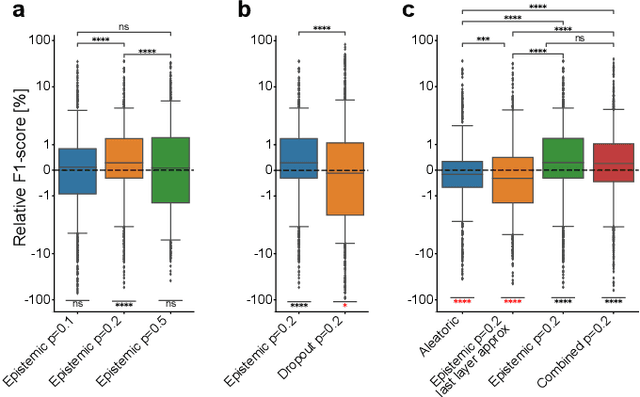
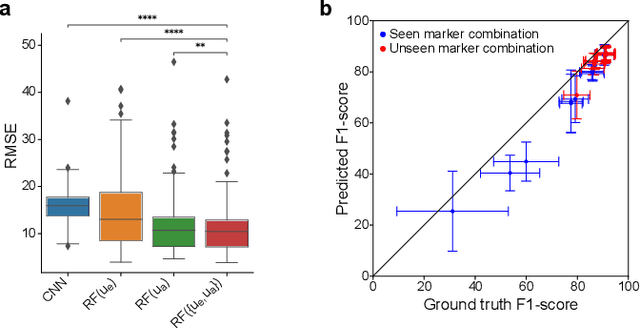
Abstract:Fluorescence microscopy images contain several channels, each indicating a marker staining the sample. Since many different marker combinations are utilized in practice, it has been challenging to apply deep learning based segmentation models, which expect a predefined channel combination for all training samples as well as at inference for future application. Recent work circumvents this problem using a modality attention approach to be effective across any possible marker combination. However, for combinations that do not exist in a labeled training dataset, one cannot have any estimation of potential segmentation quality if that combination is encountered during inference. Without this, not only one lacks quality assurance but one also does not know where to put any additional imaging and labeling effort. We herein propose a method to estimate segmentation quality on unlabeled images by (i) estimating both aleatoric and epistemic uncertainties of convolutional neural networks for image segmentation, and (ii) training a Random Forest model for the interpretation of uncertainty features via regression to their corresponding segmentation metrics. Additionally, we demonstrate that including these uncertainty measures during training can provide an improvement on segmentation performance.
Learning Ultrasound Rendering from Cross-Sectional Model Slices for Simulated Training
Jan 20, 2021



Abstract:Purpose. Given the high level of expertise required for navigation and interpretation of ultrasound images, computational simulations can facilitate the training of such skills in virtual reality. With ray-tracing based simulations, realistic ultrasound images can be generated. However, due to computational constraints for interactivity, image quality typically needs to be compromised. Methods. We propose herein to bypass any rendering and simulation process at interactive time, by conducting such simulations during a non-time-critical offline stage and then learning image translation from cross-sectional model slices to such simulated frames. We use a generative adversarial framework with a dedicated generator architecture and input feeding scheme, which both substantially improve image quality without increase in network parameters. Integral attenuation maps derived from cross-sectional model slices, texture-friendly strided convolutions, providing stochastic noise and input maps to intermediate layers in order to preserve locality are all shown herein to greatly facilitate such translation task. Results. Given several quality metrics, the proposed method with only tissue maps as input is shown to provide comparable or superior results to a state-of-the-art that uses additional images of low-quality ultrasound renderings. An extensive ablation study shows the need and benefits from the individual contributions utilized in this work, based on qualitative examples and quantitative ultrasound similarity metrics. To that end, a local histogram statistics based error metric is proposed and demonstrated for visualization of local dissimilarities between ultrasound images.
Modality Attention and Sampling Enables Deep Learning with Heterogeneous Marker Combinations in Fluorescence Microscopy
Aug 27, 2020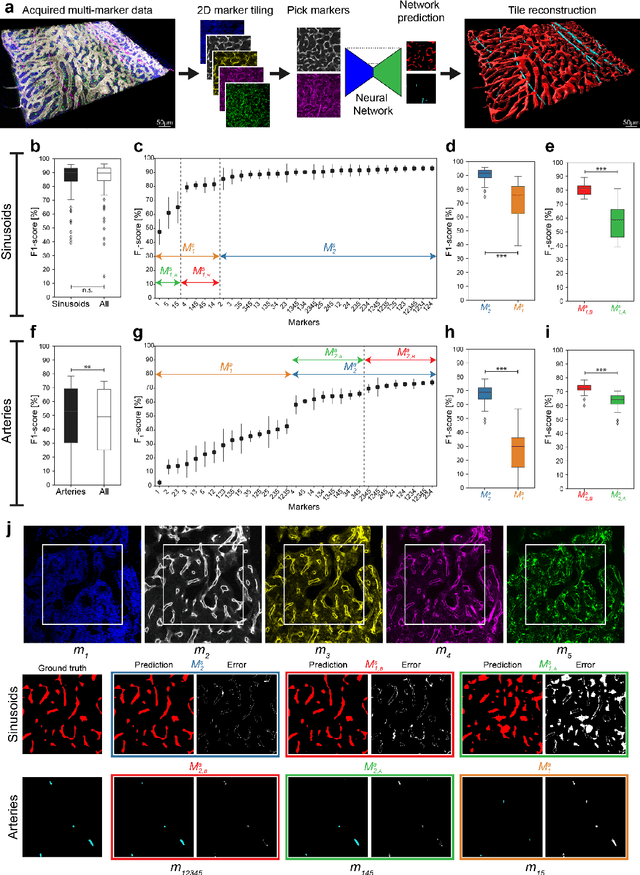

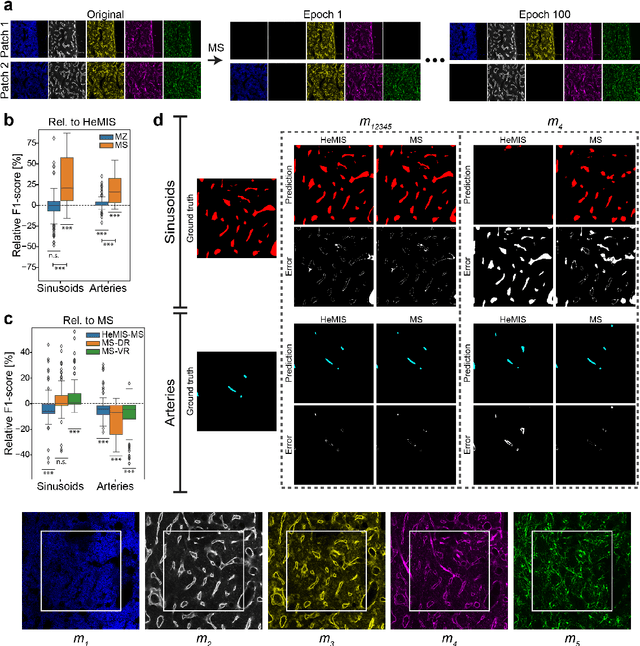
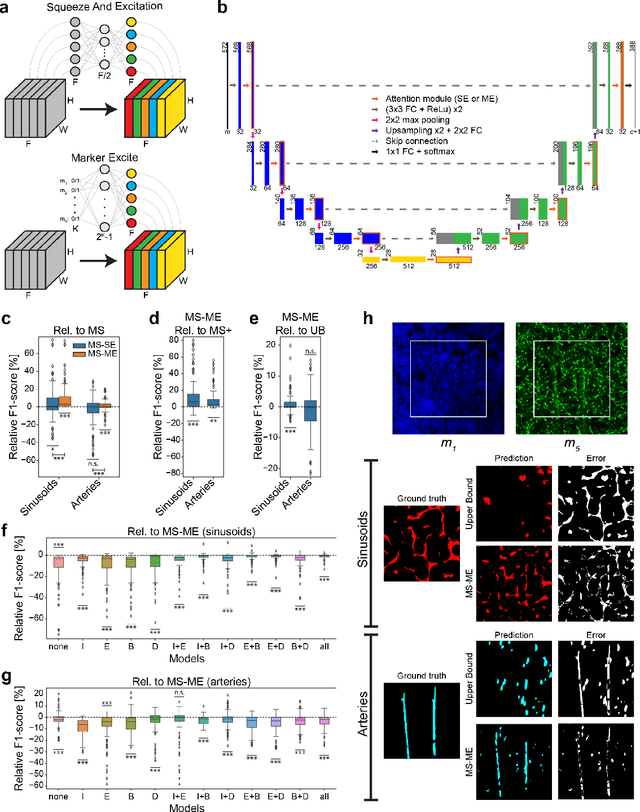
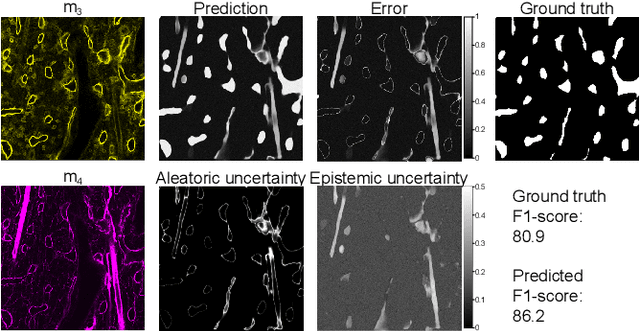
Abstract:Fluorescence microscopy allows for a detailed inspection of cells, cellular networks, and anatomical landmarks by staining with a variety of carefully-selected markers visualized as color channels. Quantitative characterization of structures in acquired images often relies on automatic image analysis methods. Despite the success of deep learning methods in other vision applications, their potential for fluorescence image analysis remains underexploited. One reason lies in the considerable workload required to train accurate models, which are normally specific for a given combination of markers, and therefore applicable to a very restricted number of experimental settings. We herein propose Marker Sampling and Excite, a neural network approach with a modality sampling strategy and a novel attention module that together enable ($i$) flexible training with heterogeneous datasets with combinations of markers and ($ii$) successful utility of learned models on arbitrary subsets of markers prospectively. We show that our single neural network solution performs comparably to an upper bound scenario where an ensemble of many networks is na\"ively trained for each possible marker combination separately. In addition, we demonstrate the feasibility of our framework in high-throughput biological analysis by revising a recent quantitative characterization of bone marrow vasculature in 3D confocal microscopy datasets. Not only can our work substantially ameliorate the use of deep learning in fluorescence microscopy analysis, but it can also be utilized in other fields with incomplete data acquisitions and missing modalities.
GramGAN: Deep 3D Texture Synthesis From 2D Exemplars
Jun 30, 2020



Abstract:We present a novel texture synthesis framework, enabling the generation of infinite, high-quality 3D textures given a 2D exemplar image. Inspired by recent advances in natural texture synthesis, we train deep neural models to generate textures by non-linearly combining learned noise frequencies. To achieve a highly realistic output conditioned on an exemplar patch, we propose a novel loss function that combines ideas from both style transfer and generative adversarial networks. In particular, we train the synthesis network to match the Gram matrices of deep features from a discriminator network. In addition, we propose two architectural concepts and an extrapolation strategy that significantly improve generalization performance. In particular, we inject both model input and condition into hidden network layers by learning to scale and bias hidden activations. Quantitative and qualitative evaluations on a diverse set of exemplars motivate our design decisions and show that our system performs superior to previous state of the art. Finally, we conduct a user study that confirms the benefits of our framework.
Deep Image Translation for Enhancing Simulated Ultrasound Images
Jun 18, 2020



Abstract:Ultrasound simulation based on ray tracing enables the synthesis of highly realistic images. It can provide an interactive environment for training sonographers as an educational tool. However, due to high computational demand, there is a trade-off between image quality and interactivity, potentially leading to sub-optimal results at interactive rates. In this work we introduce a deep learning approach based on adversarial training that mitigates this trade-off by improving the quality of simulated images with constant computation time. An image-to-image translation framework is utilized to translate low quality images into high quality versions. To incorporate anatomical information potentially lost in low quality images, we additionally provide segmentation maps to image translation. Furthermore, we propose to leverage information from acoustic attenuation maps to better preserve acoustic shadows and directional artifacts, an invaluable feature for ultrasound image interpretation. The proposed method yields an improvement of 7.2% in Fr\'{e}chet Inception Distance and 8.9% in patch-based Kullback-Leibler divergence.
Learning Generative Models using Denoising Density Estimators
Jan 08, 2020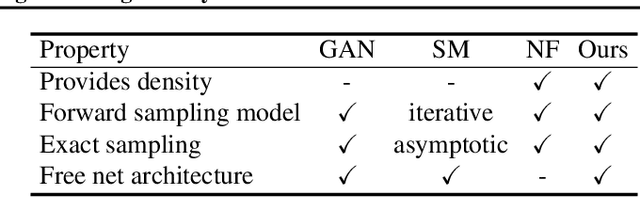
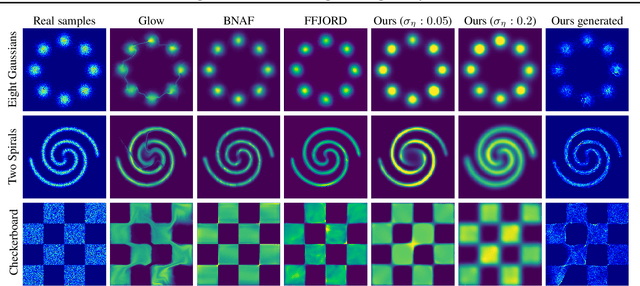
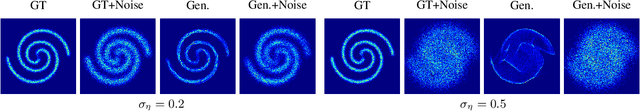
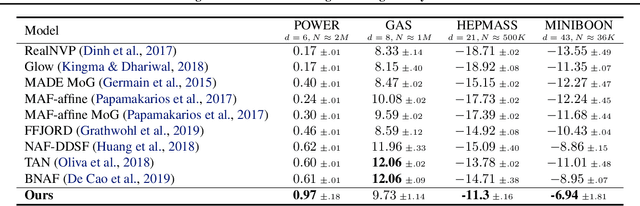
Abstract:Learning generative probabilistic models that can estimate the continuous density given a set of samples, and that can sample from that density, is one of the fundamental challenges in unsupervised machine learning. In this paper we introduce a new approach to obtain such models based on what we call denoising density estimators (DDEs). A DDE is a scalar function, parameterized by a neural network, that is efficiently trained to represent a kernel density estimator of the data. Leveraging DDEs, our main contribution is to develop a novel approach to obtain generative models that sample from given densities. We prove that our algorithms to obtain both DDEs and generative models are guaranteed to converge to the correct solutions. Advantages of our approach include that we do not require specific network architectures like in normalizing flows, ordinary differential equation solvers as in continuous normalizing flows, nor do we require adversarial training as in generative adversarial networks (GANs). Finally, we provide experimental results that demonstrate practical applications of our technique.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge