Probabilistic Spatial Analysis in Quantitative Microscopy with Uncertainty-Aware Cell Detection using Deep Bayesian Regression of Density Maps
Paper and Code
Feb 23, 2021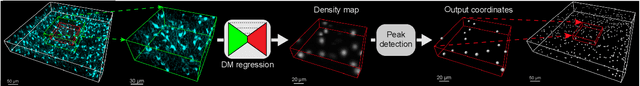

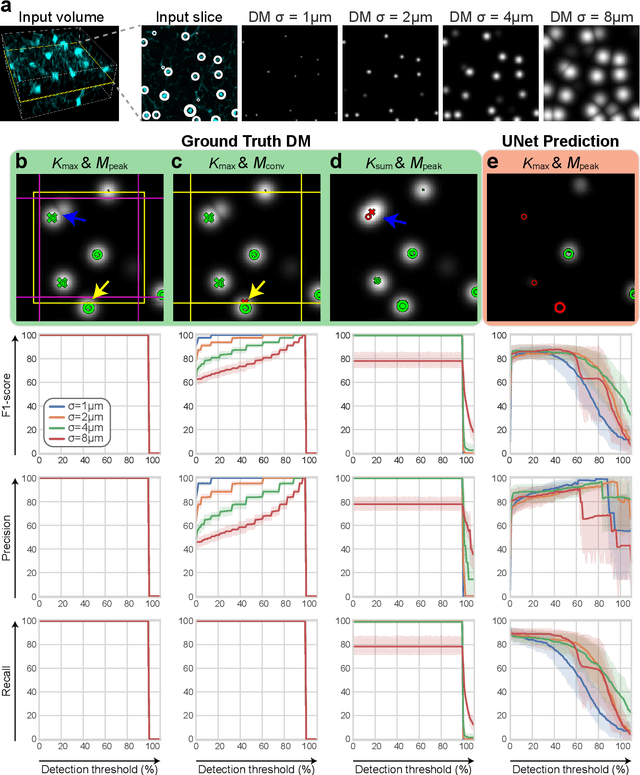
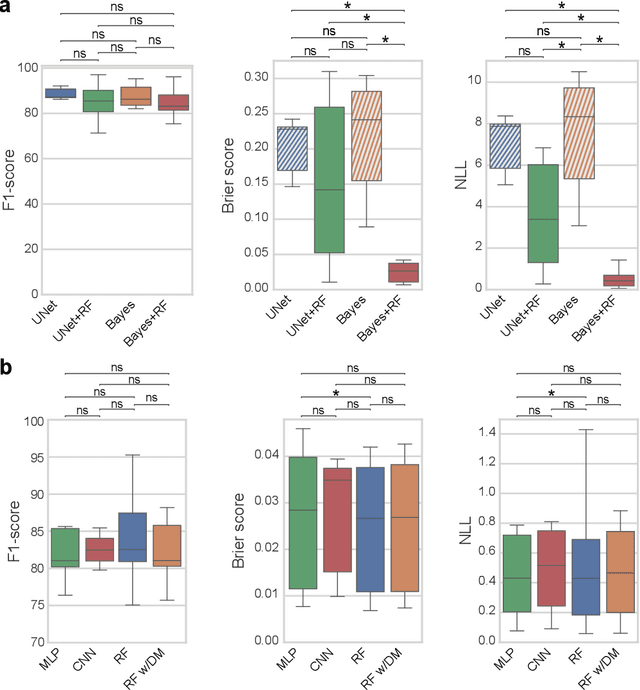
3D microscopy is key in the investigation of diverse biological systems, and the ever increasing availability of large datasets demands automatic cell identification methods that not only are accurate, but also can imply the uncertainty in their predictions to inform about potential errors and hence confidence in conclusions using them. While conventional deep learning methods often yield deterministic results, advances in deep Bayesian learning allow for accurate predictions with a probabilistic interpretation in numerous image classification and segmentation tasks. It is however nontrivial to extend such Bayesian methods to cell detection, which requires specialized learning frameworks. In particular, regression of density maps is a popular successful approach for extracting cell coordinates from local peaks in a postprocessing step, which hinders any meaningful probabilistic output. We herein propose a deep learning-based cell detection framework that can operate on large microscopy images and outputs desired probabilistic predictions by (i) integrating Bayesian techniques for the regression of uncertainty-aware density maps, where peak detection can be applied to generate cell proposals, and (ii) learning a mapping from the numerous proposals to a probabilistic space that is calibrated, i.e. accurately represents the chances of a successful prediction. Utilizing such calibrated predictions, we propose a probabilistic spatial analysis with Monte-Carlo sampling. We demonstrate this in revising an existing description of the distribution of a mesenchymal stromal cell type within the bone marrow, where our proposed methods allow us to reveal spatial patterns that are otherwise undetectable. Introducing such probabilistic analysis in quantitative microscopy pipelines will allow for reporting confidence intervals for testing biological hypotheses of spatial distributions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge